2023-07-27 カリフォルニア大学サンディエゴ校(UCSD)
◆この手法はスケーラビリティが高く、より少ないエラーや不正確性を含みます。また、既存の系統樹を逐次的に更新できる利点もあります。これにより、マイクロバイオームの研究などの分野でより迅速な解析が可能となります。将来的には、機械学習を導入することで更なる精度向上やリソースの削減が期待されています。
<関連情報>
- https://today.ucsd.edu/story/a-scalable-approach-to-building-and-updating-phylogenies
- https://www.nature.com/articles/s41587-023-01868-8
uDanceによる正確で拡張可能な系統樹の作成 Generation of accurate, expandable phylogenomic trees with uDance
Metin Balaban,Yueyu Jiang,Qiyun Zhu,Daniel McDonald,Rob Knight & Siavash Mirarab
Nature Biotechnology Published:27 July 2023
DOI:https://doi.org/10.1038/s41587-023-01868-8
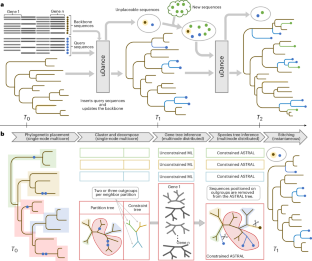
Abstract
Phylogenetic trees provide a framework for organizing evolutionary histories across the tree of life and aid downstream comparative analyses such as metagenomic identification. Methods that rely on single-marker genes such as 16S rRNA have produced trees of limited accuracy with hundreds of thousands of organisms, whereas methods that use genome-wide data are not scalable to large numbers of genomes. We introduce updating trees using divide-and-conquer (uDance), a method that enables updatable genome-wide inference using a divide-and-conquer strategy that refines different parts of the tree independently and can build off of existing trees, with high accuracy and scalability. With uDance, we infer a species tree of roughly 200,000 genomes using 387 marker genes, totaling 42.5 billion amino acid residues.

