2025-12-10 国立遺伝学研究所
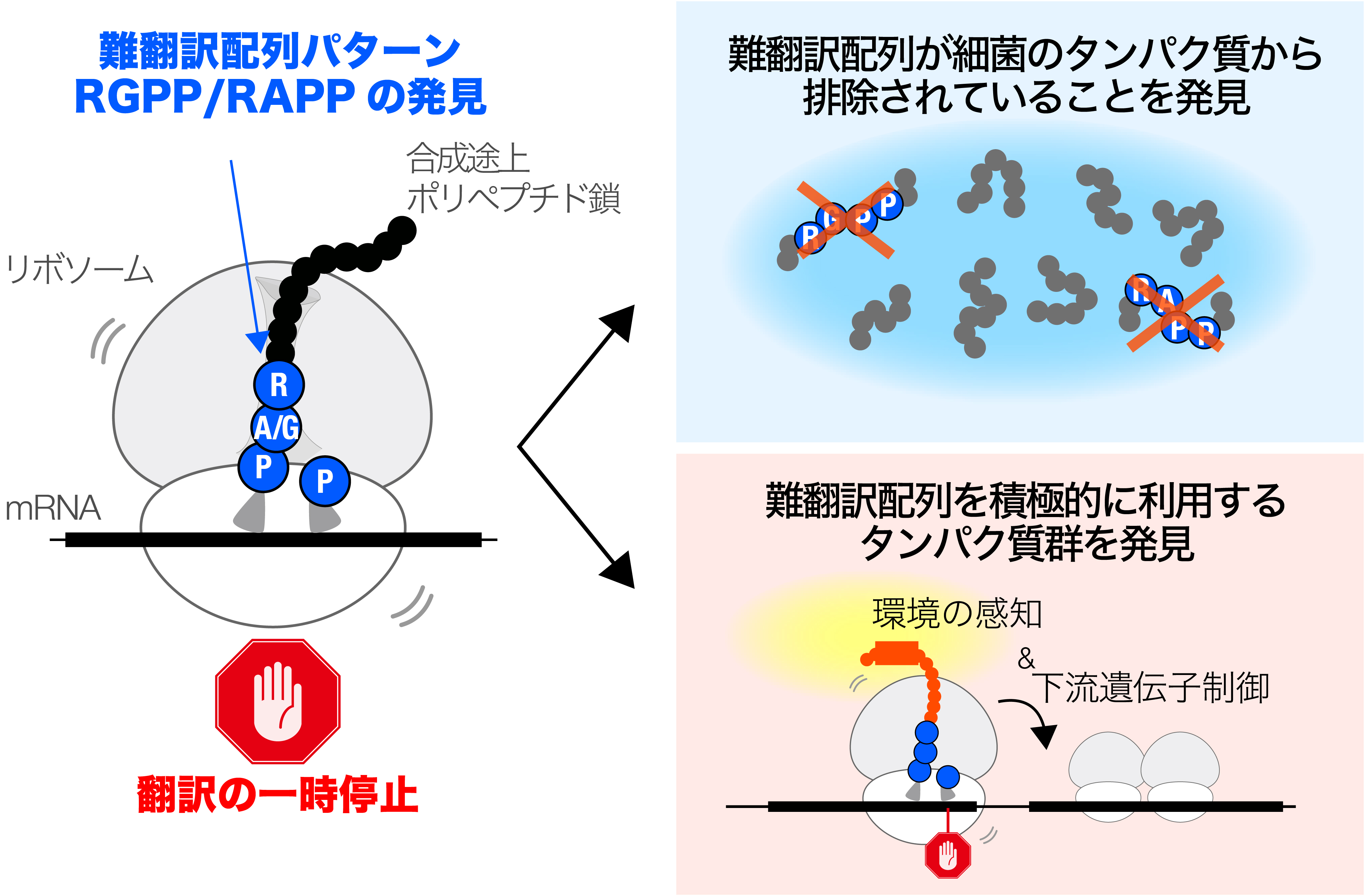
図:多くの細菌において「タンパク質の合成を止めてしまうアミノ酸配列」のパターンを発見(左)。そのような配列パターンは、一般的には、進化の過程で排除されるが(右上)、一方で、細菌は、合成困難性を細胞の機能維持に役立てるユニークなしくみを進化させることもある。
<関連情報>
- https://www.nig.ac.jp/nig/ja/2025/12/research-highlights_ja/pr20251210.html
- https://www.nig.ac.jp/nig/images/research_highlights/RH20251210.pdf
- https://link.springer.com/article/10.1038/s44318-025-00651-6
難翻訳配列に対するバクテリアプロテオームの進化的適応 Evolutionary adaptation of bacterial proteomes to translation-impeding sequences
Keigo Fujiwara,Naoko Tsuji,Karen Sakiyama,Hironori Niki & Shinobu Chiba
The EMBO Journal Published:09 December 2025
DOI:https://doi.org/10.1038/s44318-025-00651-6
Abstract
Microbial translation arrest peptides monitor intracellular environments and feedback-regulate downstream gene expression. Previous studies have identified a class of bacterial arrest peptides with C-terminal RAPP-like sequences, encoded upstream of genes involved in protein localization. In this study, we found that among RAPP-like sequences, RAPP (Arg-Ala-Pro-Pro) and RGPP (Arg-Gly-Pro-Pro) could more readily evolve into translation-impeding sequences with a particularly robust arrest that is refractory to EF-P. RAPP-like motifs were found to be strongly excluded from bacterial proteomes, likely reflecting the risk of disrupting the cellular translation system. Meanwhile, these motifs tended to occur near the C-terminus of relatively small secretory and membrane proteins. Notably, they were encoded upstream of genes with diverse functions beyond protein localization. Indeed, we identified seven RAPP/RGPP-containing arrest peptides from Streptomyces lividans encoded upstream of genes with diverse functions. These findings illustrate the bidirectional evolution of RAPP-containing proteins: their elimination from bacterial proteomes and their adaptation into arrest peptides with various regulatory roles.