2025-09-29 中国科学院(CAS)
Discovery of TranC systems reveals the molecular mechanism of CRISPR origin (Image by IGDB)
<関連情報>
- https://english.cas.cn/newsroom/research_news/life/202510/t20251009_1075519.shtml
- https://www.cell.com/cell/abstract/S0092-8674(25)01035-9
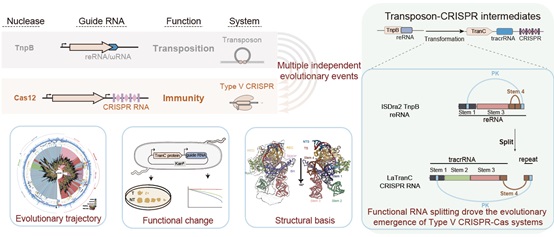
機能的なRNA分割は、トランスポゾンからV型CRISPR-Casシステムの進化的出現を促した Functional RNA splitting drove the evolutionary emergence of type V CRISPR-Cas systems from transposons
Shuai Jin ∙ Zixu Zhu ∙ Yunjia Li ∙ … ∙ Yong E. Zhang ∙ Jun-Jie Gogo Liu ∙ Caixia Gao
Cell Published:September 29, 2025
DOI:https://doi.org/10.1016/j.cell.2025.09.004
Highlights
- TranC systems are evolutionary intermediates between transposons and Cas12 effectors
- TranCs mediate DNA cleavage guided by both reRNAs and CRISPR RNAs
- Cryo-EM structures reveal a split guide RNA architecture composed of tracrRNA and crRNA
- Functional RNA splitting represents a key step in the evolution of type V CRISPR immunity
Summary
Transposon-encoded TnpB nucleases gave rise to type V CRISPR-Cas12 effectors through multiple independent domestication events. These systems use different RNA molecules as guides for DNA targeting: transposon-derived right-end RNAs (reRNAs or omega RNAs) for TnpB and CRISPR RNAs for type V CRISPR-Cas systems. However, the molecular mechanisms bridging transposon activity and CRISPR immunity remain unclear. We identify TranCs (transposon-CRISPR intermediates) derived from distinct IS605- or IS607-TnpB lineages. TranCs utilize both CRISPR RNAs and reRNAs to direct DNA cleavage. The cryoelectron microscopy (cryo-EM) structure of LaTranC from Lawsonibacter sp. closely resembles that of the ISDra2 TnpB complex; however, unlike a single-molecule reRNA, the LaTranC guide RNA is functionally split into a tracrRNA and crRNA. An engineered RNA split of ISDra2 TnpB enabled activity with a CRISPR array. These findings indicate that functional RNA splitting was the primary molecular event driving the emergence of diverse type V CRISPR-Cas systems from transposons.