2023-01-26 ペンシルベニア州立大学(PennState)
◆喫煙は、心血管疾患、がん、呼吸器疾患のリスクファクターであり、米国では毎年50万人近くが死亡している。喫煙行動は、学習によって身につけることができますが、遺伝もまた、喫煙行動をとる人のリスクに関与しています。研究者らは先行研究で、特定の遺伝子を持つ人はタバコ中毒になりやすいことを発見しています。
◆公衆衛生科学と生化学・分子生物学の教授であるDajiang Liu博士と公衆衛生科学の助教授であるBibo Jiang博士は、130万人以上の遺伝子データを用いて、機械学習を用いてこれらの大規模データセット(人の遺伝に関する特定のデータと自己申告の喫煙行動を含む)を研究する、複数の機関による大規模研究を共同主導した。
◆研究者らは、喫煙行動に関連する400以上の遺伝子を特定しました。人は何千もの遺伝子をもっている可能性があるため、そのうちのいくつかが喫煙行動に関係している理由を明らかにする必要があった。ニコチン受容体の産生を指示する遺伝子や、人をリラックスさせたり幸福感を与えたりするホルモンであるドーパミンのシグナル伝達に関与する遺伝子は、わかりやすい関連性を持っていた。残りの遺伝子については、研究チームは、それぞれが生物学的経路においてどのような役割を果たしているかを調べ、その情報をもとに、既存の経路を変更するためにすでに承認されている薬剤を割り出す必要があった。
◆この研究で得られた遺伝子データのほとんどは、ヨーロッパ系の人々のものである。そのため、機械学習モデルは、このデータだけでなく、アジア系、アフリカ系、アメリカ系の人々を含む約15万人の小規模なデータセットにも適合するように調整する必要があった。
◆Liu氏とJiang氏は、このプロジェクトで70人以上の科学者と協力しました。彼らは、風邪やインフルエンザによる咳の治療によく使われるデキストロメトルファンや、アルツハイマー病の治療に使われるガランタミンなど、禁煙に再利用できる可能性のある医薬品を少なくとも8種類特定したのです。この研究は、1月26日付のNature Geneticsに掲載されました。
<関連情報>
- https://www.psu.edu/news/research/story/machine-learning-identifies-drugs-could-potentially-help-smokers-quit/
- https://www.nature.com/articles/s41588-022-01282-x
- https://www.nature.com/articles/s41586-022-05477-4
多系統トランスクリプトームワイド関連解析により、タバコ使用の生物学と薬物再利用に関する知見が得られる Multi-ancestry transcriptome-wide association analyses yield insights into tobacco use biology and drug repurposing
Fang Chen,Xingyan Wang,Seon-Kyeong Jang,Bryan C. Quach,J. Dylan Weissenkampen,Chachrit Khunsriraksakul,Lina Yang,Renan Sauteraud,Christine M. Albert,Nicholette D. D. Allred,Donna K. Arnett,Allison E. Ashley-Koch,Kathleen C. Barnes,R. Graham Barr,Diane M. Becker,Lawrence F. Bielak,Joshua C. Bis,John Blangero,Meher Preethi Boorgula,Daniel I. Chasman,Sameer Chavan,Yii-Der I. Chen,Lee-Ming Chuang,Adolfo Correa,Joanne E. Curran,Sean P. David,Lisa de las Fuentes,Ranjan Deka,Ravindranath Duggirala,Jessica D. Faul,Melanie E. Garrett,Sina A. Gharib,Xiuqing Guo,Michael E. Hall,Nicola L. Hawley,Jiang He,Brian D. Hobbs,John E. Hokanson,Chao A. Hsiung,Shih-Jen Hwang,Thomas M. Hyde,Marguerite R. Irvin,Andrew E. Jaffe,Eric O. Johnson,Robert Kaplan,Sharon L. R. Kardia,Joel D. Kaufman,Tanika N. Kelly,Joel E. Kleinman,Charles Kooperberg,I-Te Lee,Daniel Levy,Sharon M. Lutz,Ani W. Manichaikul,Lisa W. Martin,Olivia Marx,Stephen T. McGarvey,Ryan L. Minster,Matthew Moll,Karine A. Moussa,Take Naseri,Kari E. North,Elizabeth C. Oelsner,Juan M. Peralta,Patricia A. Peyser,Bruce M. Psaty,Nicholas Rafaels,Laura M. Raffield,Muagututi’a Sefuiva Reupena,Stephen S. Rich,Jerome I. Rotter,David A. Schwartz,Aladdin H. Shadyab,Wayne H-H. Sheu,Mario Sims,Jennifer A. Smith,Xiao Sun,Kent D. Taylor,Marilyn J. Telen,Harold Watson,Daniel E. Weeks,David R. Weir,Lisa R. Yanek,Kendra A. Young,Kristin L. Young,Wei Zhao,Dana B. Hancock,Bibo Jiang,Scott Vrieze & Dajiang J. Liu
Nature Genetics Published:26 January 2023
DOI:https://doi.org/10.1038/s41588-022-01282-x
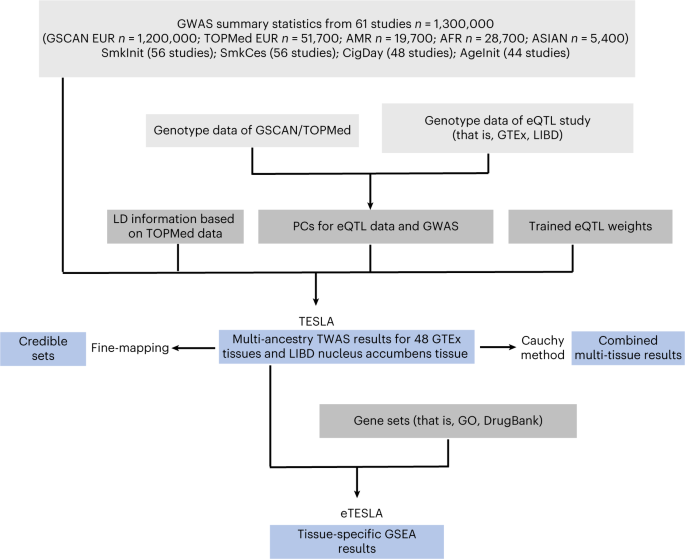
Abstract
Most transcriptome-wide association studies (TWASs) so far focus on European ancestry and lack diversity. To overcome this limitation, we aggregated genome-wide association study (GWAS) summary statistics, whole-genome sequences and expression quantitative trait locus (eQTL) data from diverse ancestries. We developed a new approach, TESLA (multi-ancestry integrative study using an optimal linear combination of association statistics), to integrate an eQTL dataset with a multi-ancestry GWAS. By exploiting shared phenotypic effects between ancestries and accommodating potential effect heterogeneities, TESLA improves power over other TWAS methods. When applied to tobacco use phenotypes, TESLA identified 273 new genes, up to 55% more compared with alternative TWAS methods. These hits and subsequent fine mapping using TESLA point to target genes with biological relevance. In silico drug-repurposing analyses highlight several drugs with known efficacy, including dextromethorphan and galantamine, and new drugs such as muscle relaxants that may be repurposed for treating nicotine addiction.
遺伝子の多様性がタバコやアルコールの遺伝子発見を促進する Genetic diversity fuels gene discovery for tobacco and alcohol use
Gretchen R. B. Saunders,Xingyan Wang,Fang Chen,Seon-Kyeong Jang,Mengzhen Liu,Chen Wang,Shuang Gao,Yu Jiang,Chachrit Khunsriraksakul,Jacqueline M. Otto,Clifton Addison,Masato Akiyama,Christine M. Albert,Fazil Aliev,Alvaro Alonso,Donna K. Arnett,Allison E. Ashley-Koch,Aneel A. Ashrani,Kathleen C. Barnes,R. Graham Barr,Traci M. Bartz,Diane M. Becker,Lawrence F. Bielak,Emelia J. Benjamin,Joshua C. Bis,Gyda Bjornsdottir,John Blangero,Eugene R. Bleecker,Jason D. Boardman,Eric Boerwinkle,Dorret I. Boomsma,Meher Preethi Boorgula,Donald W. Bowden,Jennifer A. Brody,Brian E. Cade,Daniel I. Chasman,Sameer Chavan,Yii-Der Ida Chen,Zhengming Chen,Iona Cheng,Michael H. Cho,Hélène Choquet,John W. Cole,Marilyn C. Cornelis,Francesco Cucca,Joanne E. Curran,Mariza de Andrade,Danielle M. Dick,Anna R. Docherty,Ravindranath Duggirala,Charles B. Eaton,Marissa A. Ehringer,Tõnu Esko,Jessica D. Faul,Lilian Fernandes Silva,Edoardo Fiorillo,Myriam Fornage,Barry I. Freedman,Maiken E. Gabrielsen,Melanie E. Garrett,Sina A. Gharib,Christian Gieger,Nathan Gillespie,David C. Glahn,Scott D. Gordon,Charles C. Gu,Dongfeng Gu,Daniel F. Gudbjartsson,Xiuqing Guo,Jeffrey Haessler,Michael E. Hall,Toomas Haller,Kathleen Mullan Harris,Jiang He,Pamela Herd,John K. Hewitt,Ian Hickie,Bertha Hidalgo,John E. Hokanson,Christian Hopfer,JoukeJan Hottenga,Lifang Hou,Hongyan Huang,Yi-Jen Hung,David J. Hunter,Kristian Hveem,Shih-Jen Hwang,Chii-Min Hwu,William Iacono,Marguerite R. Irvin,Yon Ho Jee,Eric O. Johnson,Yoonjung Y. Joo,Eric Jorgenson,Anne E. Justice,Yoichiro Kamatani,Robert C. Kaplan,Jaakko Kaprio,Sharon L. R. Kardia,Matthew C. Keller,Tanika N. Kelly,Charles Kooperberg,Tellervo Korhonen,Peter Kraft,Kenneth Krauter,Johanna Kuusisto,Markku Laakso,Jessica Lasky-Su,Wen-Jane Lee,James J. Lee,Daniel Levy,Liming Li,Kevin Li,Yuqing Li,Kuang Lin,Penelope A. Lind,Chunyu Liu,Donald M. Lloyd-Jones,Sharon M. Lutz,Jiantao Ma,Reedik Mägi,Ani Manichaikul,Nicholas G. Martin,Ravi Mathur,Nana Matoba,Patrick F. McArdle,Matt McGue,Matthew B. McQueen,Sarah E. Medland,Andres Metspalu,Deborah A. Meyers,Iona Y. Millwood,Braxton D. Mitchell,Karen L. Mohlke,Matthew Moll,May E. Montasser,Alanna C. Morrison,Antonella Mulas,Jonas B. Nielsen,Kari E. North,Elizabeth C. Oelsner,Yukinori Okada,Valeria Orrù,Nicholette D. Palmer,Teemu Palviainen,Anita Pandit,S. Lani Park,Ulrike Peters,Annette Peters,Patricia A. Peyser,Tinca J. C. Polderman,Nicholas Rafaels,Susan Redline,Robert M. Reed,Alex P. Reiner,John P. Rice,Stephen S. Rich,Nicole E. Richmond,Carol Roan,Jerome I. Rotter,Michael N. Rueschman,Valgerdur Runarsdottir,Nancy L. Saccone,David A. Schwartz,Aladdin H. Shadyab,Jingchunzi Shi,Suyash S. Shringarpure,Kamil Sicinski,Anne Heidi Skogholt,Jennifer A. Smith,Nicholas L. Smith,Nona Sotoodehnia,Michael C. Stallings,Hreinn Stefansson,Kari Stefansson,Jerry A. Stitzel,Xiao Sun,Moin Syed,Ruth Tal-Singer,Amy E. Taylor,Kent D. Taylor,Marilyn J. Telen,Khanh K. Thai,Hemant Tiwari,Constance Turman,Thorarinn Tyrfingsson,Tamara L. Wall,Robin G. Walters,David R. Weir,Scott T. Weiss,Wendy B. White,John B. Whitfield,Kerri L. Wiggins,Gonneke Willemsen,Cristen J. Willer,Bendik S. Winsvold,Huichun Xu,Lisa R. Yanek,Jie Yin,Kristin L. Young,Kendra A. Young,Bing Yu,Wei Zhao,Wei Zhou,Sebastian Zöllner,Luisa Zuccolo,23andMe Research Team,The Biobank Japan Project,Chiara Batini,Andrew W. Bergen,Laura J. Bierut,Sean P. David,Sarah A. Gagliano Taliun,Dana B. Hancock,Bibo Jiang,Marcus R. Munafò,Thorgeir E. Thorgeirsson,Dajiang J. Liu & Scott Vrieze
Nature Published:07 December 2022
DOI:https://doi.org/10.1038/s41586-022-05477-4
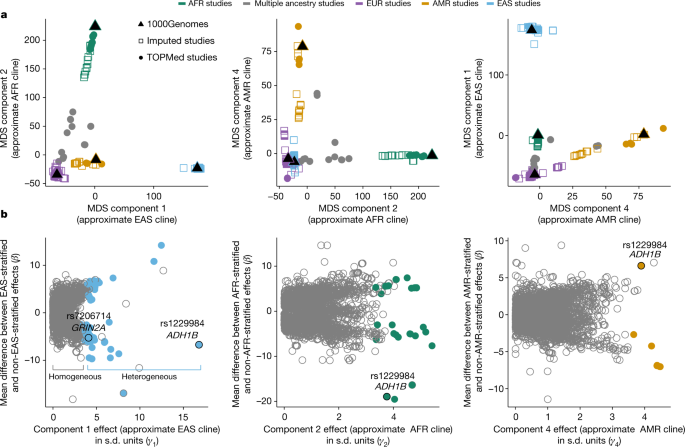
Abstract
Tobacco and alcohol use are heritable behaviours associated with 15% and 5.3% of worldwide deaths, respectively, due largely to broad increased risk for disease and injury1,2,3,4. These substances are used across the globe, yet genome-wide association studies have focused largely on individuals of European ancestries5. Here we leveraged global genetic diversity across 3.4 million individuals from four major clines of global ancestry (approximately 21% non-European) to power the discovery and fine-mapping of genomic loci associated with tobacco and alcohol use, to inform function of these loci via ancestry-aware transcriptome-wide association studies, and to evaluate the genetic architecture and predictive power of polygenic risk within and across populations. We found that increases in sample size and genetic diversity improved locus identification and fine-mapping resolution, and that a large majority of the 3,823 associated variants (from 2,143 loci) showed consistent effect sizes across ancestry dimensions. However, polygenic risk scores developed in one ancestry performed poorly in others, highlighting the continued need to increase sample sizes of diverse ancestries to realize any potential benefit of polygenic prediction.

