2023-04-06 エディンバラ大学
この研究は、英国の約4,900万人のうち90%がタイプ2糖尿病を患っているという深刻な状況に対処するためのものです。
<関連情報>
- https://www.ed.ac.uk/news/2023/new-test-could-help-identify-type-2-diabetes-risk
- https://www.nature.com/articles/s43587-023-00391-4
欧州の2つのコホートにおけるDNAメチル化スコアの開発と検証により、2型糖尿病の10年リスク予測が増強される Development and validation of DNA methylation scores in two European cohorts augment 10-year risk prediction of type 2 diabetes
Yipeng Cheng,Danni A. Gadd,Christian Gieger,Karla Monterrubio-Gómez,Yufei Zhang,Imrich Berta,Michael J. Stam,Natalia Szlachetka,Evgenii Lobzaev,Nicola Wrobel,Lee Murphy,Archie Campbell,Cliff Nangle,Rosie M. Walker,Chloe Fawns-Ritchie,Annette Peters,Wolfgang Rathmann,David J. Porteous,Kathryn L. Evans,Andrew M. McIntosh,Timothy I. Cannings,Melanie Waldenberger,Andrea Ganna,Daniel L. McCartney,Catalina A. Vallejos & Riccardo E. Marioni
Nature Aging Published:06 April 2023
DOI:https://doi.org/10.1038/s43587-023-00391-4
Abstract
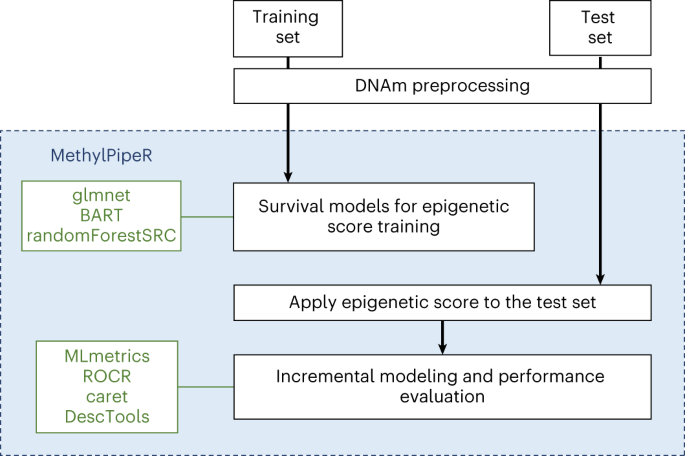
Type 2 diabetes mellitus (T2D) presents a major health and economic burden that could be alleviated with improved early prediction and intervention. While standard risk factors have shown good predictive performance, we show that the use of blood-based DNA methylation information leads to a significant improvement in the prediction of 10-year T2D incidence risk. Previous studies have been largely constrained by linear assumptions, the use of cytosine–guanine pairs one-at-a-time and binary outcomes. We present a flexible approach (via an R package, MethylPipeR) based on a range of linear and tree-ensemble models that incorporate time-to-event data for prediction. Using the Generation Scotland cohort (training set ncases = 374, ncontrols = 9,461; test set ncases = 252, ncontrols = 4,526) our best-performing model (area under the receiver operating characteristic curve (AUC) = 0.872, area under the precision-recall curve (PRAUC) = 0.302) showed notable improvement in 10-year onset prediction beyond standard risk factors (AUC = 0.839, precision–recall AUC = 0.227). Replication was observed in the German-based KORA study (n = 1,451, ncases = 142, P = 1.6 × 10-5).