2023-11-20 チャルマース工科大学
◆研究プロジェクトを率いたアンドレアス・ダーリン博士は、「当社の手法は、さまざまな疾患の早期で危険なプロセスを理解する可能性があり、最終的にはそれらに対抗する薬物の作用メカニズムについての知識をもたらす可能性があると考えています。」と述べています。新しいトラップは、従来は難しかった凝集体がまだ非常に小さい段階である初期の発展を追跡するのに役立つとされています。
<関連情報>
- https://news.cision.com/chalmers/r/tiny-traps-can-provide-new-knowledge-about-difficult-to-treat-diseases,c3875673
- https://www.nature.com/articles/s41467-023-40889-4
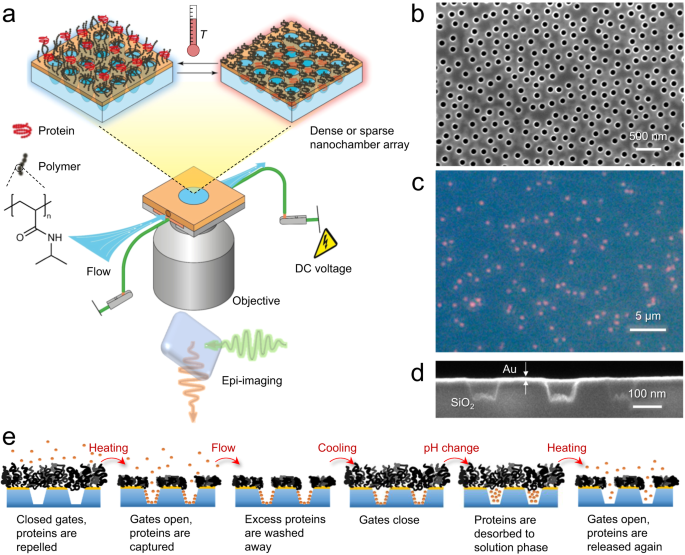
高分子ゲート付きナノスケールチャンバーを用いた生理的条件下での複数タンパク質の安定した捕捉 Stable trapping of multiple proteins at physiological conditions using nanoscale chambers with macromolecular gates
Justas Svirelis,Zeynep Adali,Gustav Emilsson,Jesper Medin,John Andersson,Radhika Vattikunta,Mats Hulander,Julia Järlebark,Krzysztof Kolman,Oliver Olsson,Yusuke Sakiyama,Roderick Y. H. Lim & Andreas Dahlin
Nature Communications Published:23 August 2023
DOI:https://doi.org/10.1038/s41467-023-40889-4
Abstract
The possibility to detect and analyze single or few biological molecules is very important for understanding interactions and reaction mechanisms. Ideally, the molecules should be confined to a nanoscale volume so that the observation time by optical methods can be extended. However, it has proven difficult to develop reliable, non-invasive trapping techniques for biomolecules under physiological conditions. Here we present a platform for long-term tether-free (solution phase) trapping of proteins without exposing them to any field gradient forces. We show that a responsive polymer brush can make solid state nanopores switch between a fully open and a fully closed state with respect to proteins, while always allowing the passage of solvent, ions and small molecules. This makes it possible to trap a very high number of proteins (500-1000) inside nanoscale chambers as small as one attoliter, reaching concentrations up to 60 gL-1. Our method is fully compatible with parallelization by imaging arrays of nanochambers. Additionally, we show that enzymatic cascade reactions can be performed with multiple native enzymes under full nanoscale confinement and steady supply of reactants. This platform will greatly extend the possibilities to optically analyze interactions involving multiple proteins, such as the dynamics of oligomerization events.