2023-11-16 マックス・プランク研究所
◆ディープラーニングに基づくアルゴリズムを用いて設計された数千の抗微生物ペプチドを、新しい細胞外蛋白質合成パイプラインで迅速かつ費用対効果の高い方法で合成。その結果、30の機能的なペプチドが同定され、そのうち6つは多剤耐性病原体に対して広範な活性を示し、細菌が耐性を形成しなかった。
◆この新手法は生体活性ペプチドの迅速で効果的な開発に貢献し、抗微生物薬の新規制御法の可能性を示唆しています。
<関連情報>
- https://www.mpg.de/21130556/1116-terr-cell-free-quest-for-new-antibiotics-153410-x?c=2249
- https://www.nature.com/articles/s41467-023-42434-9
無細胞生合成とディープラーニングの組み合わせが抗菌ペプチドの新規開発を加速する Cell-free biosynthesis combined with deep learning accelerates de novo-development of antimicrobial peptides
Amir Pandi,David Adam,Amir Zare,Van Tuan Trinh,Stefan L. Schaefer,Marie Burt,Björn Klabunde,Elizaveta Bobkova,Manish Kushwaha,Yeganeh Foroughijabbari,Peter Braun,Christoph Spahn,Christian Preußer,Elke Pogge von Strandmann,Helge B. Bode,Heiner von Buttlar,Wilhelm Bertrams,Anna Lena Jung,Frank Abendroth,Bernd Schmeck,Gerhard Hummer,Olalla Vázquez & Tobias J. Erb
Nature Communications Published:08 November 2023
DOI:https://doi.org/10.1038/s41467-023-42434-9
Abstract
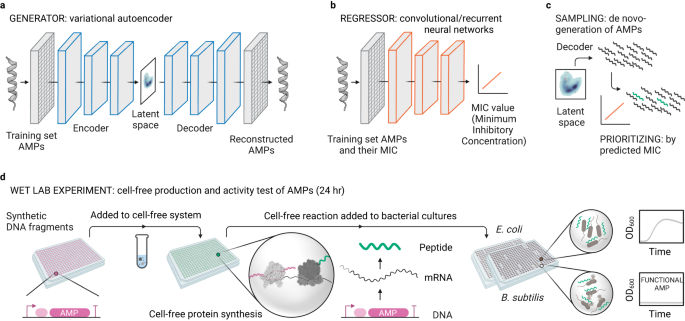
Bioactive peptides are key molecules in health and medicine. Deep learning holds a big promise for the discovery and design of bioactive peptides. Yet, suitable experimental approaches are required to validate candidates in high throughput and at low cost. Here, we established a cell-free protein synthesis (CFPS) pipeline for the rapid and inexpensive production of antimicrobial peptides (AMPs) directly from DNA templates. To validate our platform, we used deep learning to design thousands of AMPs de novo. Using computational methods, we prioritized 500 candidates that we produced and screened with our CFPS pipeline. We identified 30 functional AMPs, which we characterized further through molecular dynamics simulations, antimicrobial activity and toxicity. Notably, six de novo-AMPs feature broad-spectrum activity against multidrug-resistant pathogens and do not develop bacterial resistance. Our work demonstrates the potential of CFPS for high throughput and low-cost production and testing of bioactive peptides within less than 24 h.