2023-11-28 マサチューセッツ工科大学(MIT)
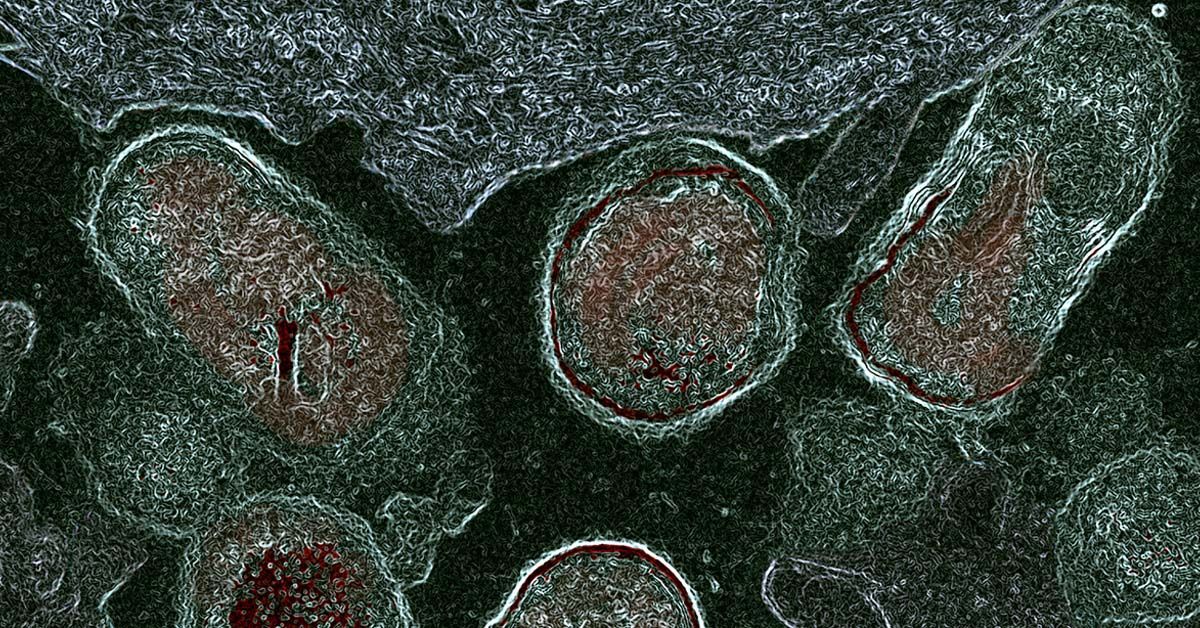
◆この手法は、緑または赤色の蛍光分子を使用し、それぞれ異なる速度で点滅する特性を利用しています。数秒、数分、または数時間にわたる細胞のイメージングを行い、計算アルゴリズムを使用して各対象タンパク質の量を時間とともに追跡できます。
◆これにより、細胞の活動や異常(老化や病気など)に関する理解が向上し、生物学の基本的な問題に取り組む手助けとなります。
<関連情報>
- https://news.mit.edu/2023/new-method-fluorescent-labels-living-cell-1128
- https://www.cell.com/cell/fulltext/S0092-8674(23)01227-8
生細胞内の動的シグナル伝達ネットワークの時間多重イメージング Temporally multiplexed imaging of dynamic signaling networks in living cells
Yong Qian,Orhan T. Celiker,Zeguan Wang,Burcu Guner-Ataman,Edward S. Boyden,
Cell Published:November 28, 2023
DOI:https://doi.org/10.1016/j.cell.2023.11.010
Highlights
•TMI computationally separates signals from fluorophores with different switching kinetics
•TMI enables imaging of many signals at once, in living cells, using standard microscopes
•TMI can be used to image cytoskeletal structures and organelles in living cells
•TMI can report gene expression, kinase activity, protein movement, and other signals
Summary
Molecular signals interact in networks to mediate biological processes. To analyze these networks, it would be useful to image many signals at once, in the same living cell, using standard microscopes and genetically encoded fluorescent reporters. Here, we report temporally multiplexed imaging (TMI), which uses genetically encoded fluorescent proteins with different clocklike properties—such as reversibly photoswitchable fluorescent proteins with different switching kinetics—to represent different cellular signals. We linearly decompose a brief (few-second-long) trace of the fluorescence fluctuations, at each point in a cell, into a weighted sum of the traces exhibited by each fluorophore expressed in the cell. The weights then represent the signal amplitudes. We use TMI to analyze relationships between different kinase activities in individual cells, as well as between different cell-cycle signals, pointing toward broad utility throughout biology in the analysis of signal transduction cascades in living systems.
Graphical abstract