<関連情報>
- https://actu.epfl.ch/news/ai-tool-maps-out-cell-metabolism-with-precision/
- https://www.nature.com/articles/s41929-024-01220-6
生成的機械学習により、細胞内の代謝状態を正確に特徴付ける動力学モデルが作成される Generative machine learning produces kinetic models that accurately characterize intracellular metabolic states
Subham Choudhury,Bharath Narayanan,Michael Moret,Vassily Hatzimanikatis & Ljubisa Miskovic
Nature Catalysis Published:30 August 2024
DOI:https://doi.org/10.1038/s41929-024-01220-6
Abstract
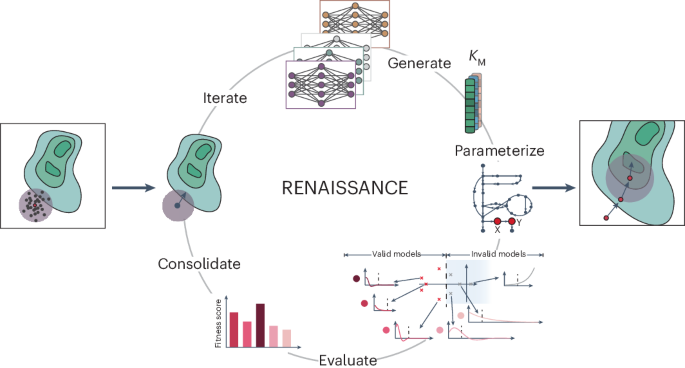
Generating large omics datasets has become routine for gaining insights into cellular processes, yet deciphering these datasets to determine metabolic states remains challenging. Kinetic models can help integrate omics data by explicitly linking metabolite concentrations, metabolic fluxes and enzyme levels. Nevertheless, determining the kinetic parameters that underlie cellular physiology poses notable obstacles to the widespread use of these mathematical representations of metabolism. Here we present RENAISSANCE, a generative machine learning framework for efficiently parameterizing large-scale kinetic models with dynamic properties matching experimental observations. Through seamless integration of diverse omics data and other relevant information, including extracellular medium composition, physicochemical data and expertise of domain specialists, RENAISSANCE accurately characterizes intracellular metabolic states in Escherichia coli. It also estimates missing kinetic parameters and reconciles them with sparse experimental data, substantially reducing parameter uncertainty and improving accuracy. This framework will be valuable for researchers studying metabolic variations involving changes in metabolite and enzyme levels and enzyme activity in health and biotechnology.