2024-10-07 コロンビア大学
<関連情報>
- https://cancerdynamics.columbia.edu/news/mapping-multimodal-phenotypes-perturbations-cells-and-tissue-crisprmap
- https://www.nature.com/articles/s41587-024-02386-x
CRISPRmapによる細胞・組織内の摂動とマルチモーダル表現型のマッピング Mapping multimodal phenotypes to perturbations in cells and tissue with CRISPRmap
Jiacheng Gu,Abhishek Iyer,Ben Wesley,Angelo Taglialatela,Giuseppe Leuzzi,Sho Hangai,Aubrianna Decker,Ruoyu Gu,Naomi Klickstein,Yuanlong Shuai,Kristina Jankovic,Lucy Parker-Burns,Yinuo Jin,Jia Yi Zhang,Justin Hong,Xiang Niu,Jonathon A. Costa,Mikael G. Pezet,Jacqueline Chou,Hans-Willem Snoeck,Dan A. Landau,Elham Azizi,Edmond M. Chan,Alberto Ciccia & Jellert T. Gaublomme
Nature Biotechnology Published:07 October 2024
DOI:https://doi.org/10.1038/s41587-024-02386-x
Abstract
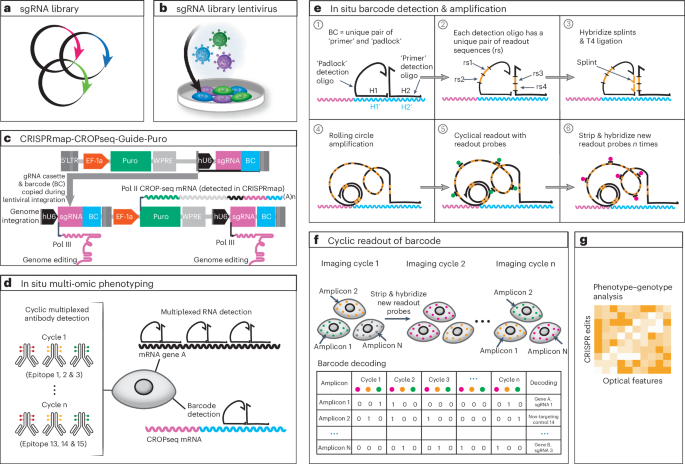
Unlike sequencing-based methods, which require cell lysis, optical pooled genetic screens enable investigation of spatial phenotypes, including cell morphology, protein subcellular localization, cell–cell interactions and tissue organization, in response to targeted CRISPR perturbations. Here we report a multimodal optical pooled CRISPR screening method, which we call CRISPRmap. CRISPRmap combines in situ CRISPR guide-identifying barcode readout with multiplexed immunofluorescence and RNA detection. Barcodes are detected and read out through combinatorial hybridization of DNA oligos, enhancing barcode detection efficiency. CRISPRmap enables in situ barcode readout in cell types and contexts that were elusive to conventional optical pooled screening, including cultured primary cells, embryonic stem cells, induced pluripotent stem cells, derived neurons and in vivo cells in a tissue context. We conducted a screen in a breast cancer cell line of the effects of DNA damage repair gene variants on cellular responses to commonly used cancer therapies, and we show that optical phenotyping pinpoints likely pathogenic patient-derived mutations that were previously classified as variants of unknown clinical significance.