2024-11-04 リンショーピング大学
<関連情報>
- https://liu.se/en/news-item/liu-forskare-far-alphafold-att-forutse-valdigt-stora-proteiner
- https://www.nature.com/articles/s41467-024-52951-w
多量体複合体における実験と予測を統合するためのAlphaFoldのマスク解除 Unmasking AlphaFold to integrate experiments and predictions in multimeric complexes
Claudio Mirabello,Björn Wallner,Björn Nystedt,Stavros Azinas & Marta Carroni
Nature Communications Published:09 October 2024
DOI:https://doi.org/10.1038/s41467-024-52951-w
Abstract
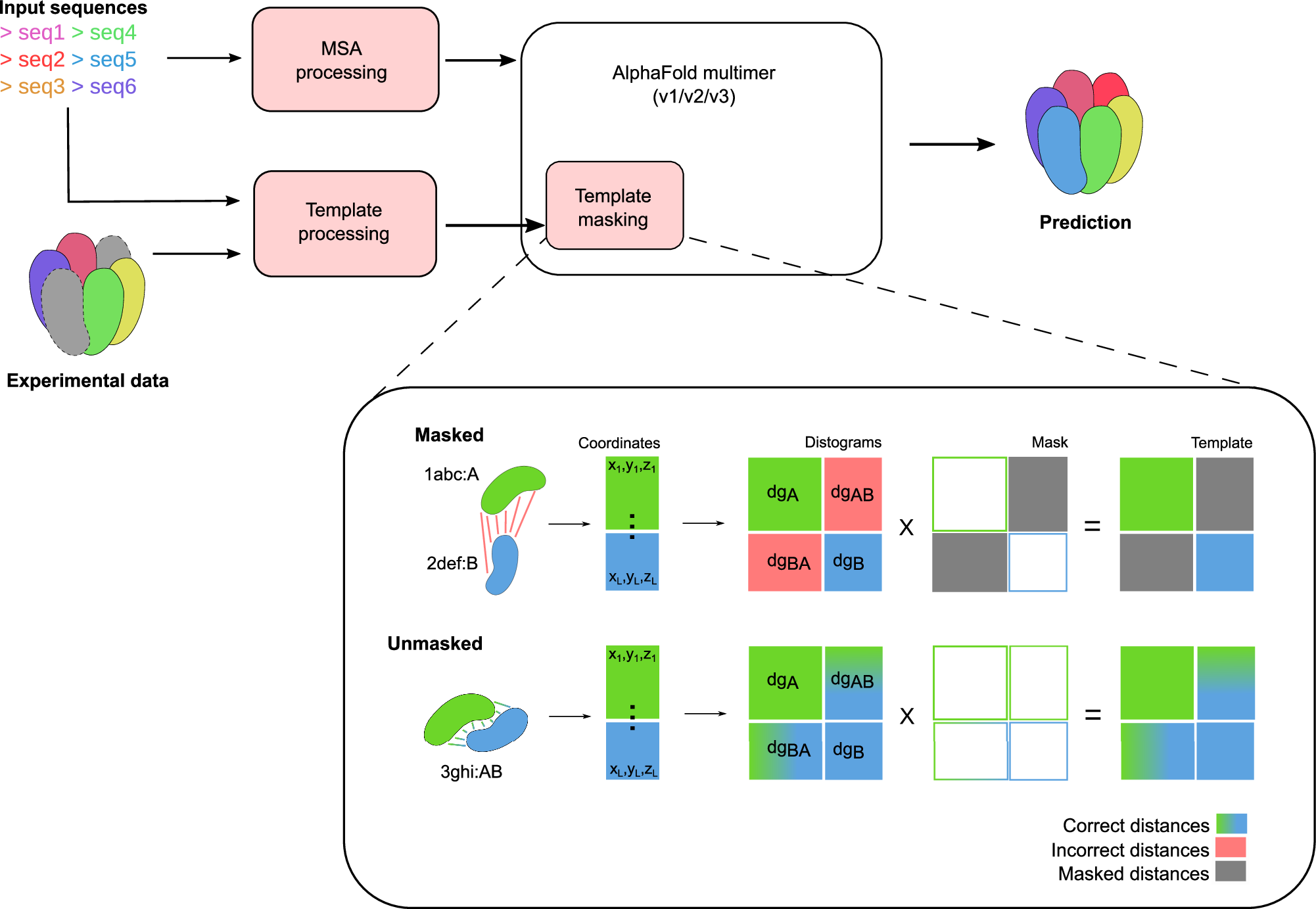
Since the release of AlphaFold, researchers have actively refined its predictions and attempted to integrate it into existing pipelines for determining protein structures. These efforts have introduced a number of functionalities and optimisations at the latest Critical Assessment of protein Structure Prediction edition (CASP15), resulting in a marked improvement in the prediction of multimeric protein structures. However, AlphaFold’s capability of predicting large protein complexes is still limited and integrating experimental data in the prediction pipeline is not straightforward. In this study, we introduce AF_unmasked to overcome these limitations. Our results demonstrate that AF_unmasked can integrate experimental information to build larger or hard to predict protein assemblies with high confidence. The resulting predictions can help interpret and augment experimental data. This approach generates high quality (DockQ score > 0.8) structures even when little to no evolutionary information is available and imperfect experimental structures are used as a starting point. AF_unmasked is developed and optimised to fill incomplete experimental structures (structural inpainting), which may provide insights into protein dynamics. In summary, AF_unmasked provides an easy-to-use method that efficiently integrates experiments to predict large protein complexes more confidently.