2024-12-04 ミュンヘン大学(LMU)
<関連情報>
- https://www.lmu.de/en/newsroom/news-overview/news/discovery-of-molecular-switch-for-protein-production.html
- https://www.nature.com/articles/s41467-024-54556-9
EF-PとそのパラログEfpL(YeiP)がプロリンを含む配列の翻訳を差次的に制御することを発見 EF-P and its paralog EfpL (YeiP) differentially control translation of proline-containing sequences
Alina Sieber,Marina Parr,Julian von Ehr,Karthikeyan Dhamotharan,Pavel Kielkowski,Tess Brewer,Anna Schäpers,Ralph Krafczyk,Fei Qi,Andreas Schlundt,Dmitrij Frishman &Jürgen Lassak
Nature Communications Published:02 December 2024
DOI:https://doi.org/10.1038/s41467-024-54556-9
Abstract
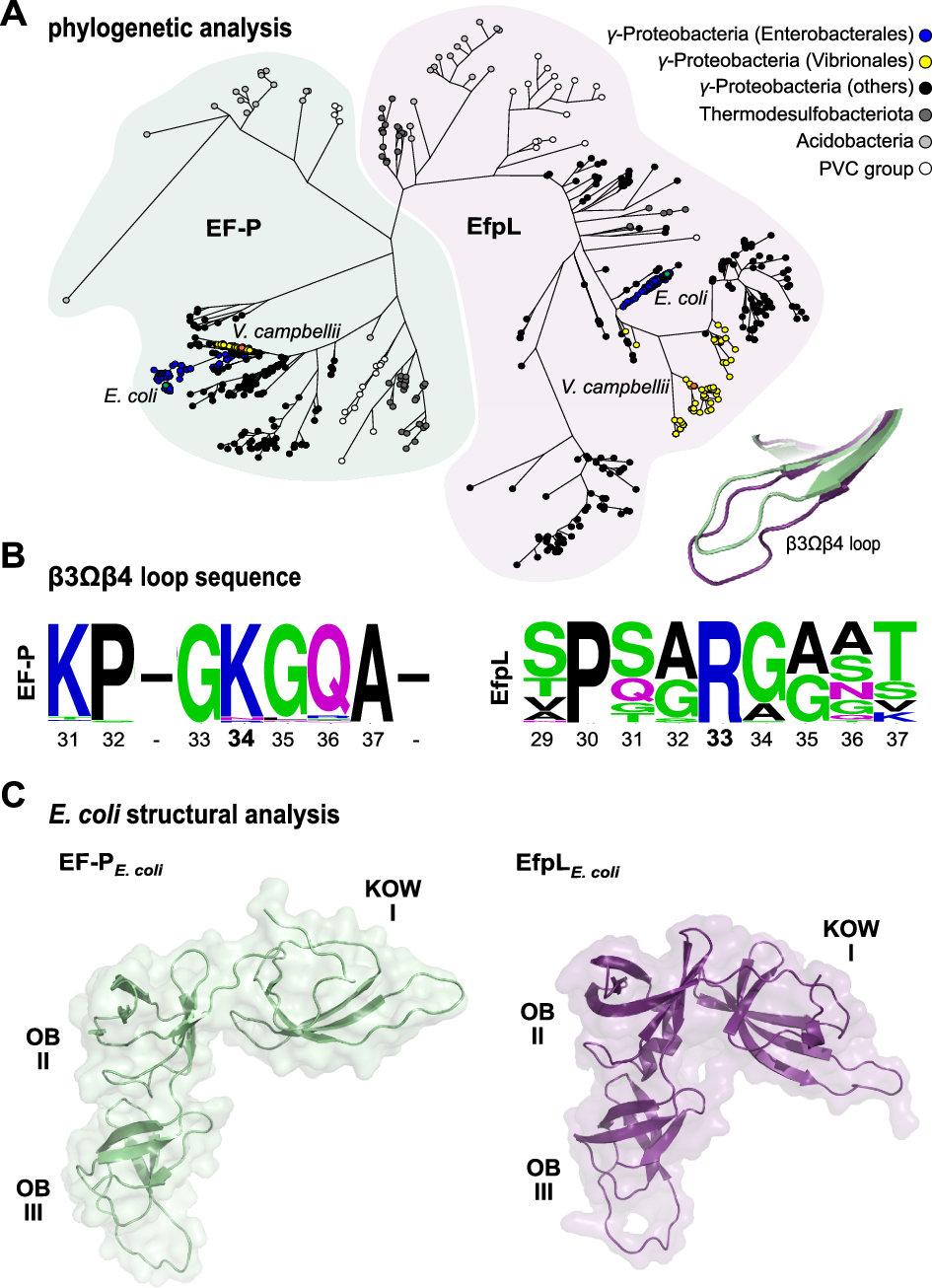
Polyproline sequences are deleterious to cells because they stall ribosomes. In bacteria, EF-P plays an important role in overcoming such polyproline sequence-induced ribosome stalling. Additionally, numerous bacteria possess an EF-P paralog called EfpL (also known as YeiP) of unknown function. Here, we functionally and structurally characterize EfpL from Escherichia coli and demonstrate its role in the translational stress response. Through ribosome profiling, we analyze the EfpL arrest motif spectrum and find additional sequences beyond the canonical polyproline motifs that both EF-P and EfpL can resolve. Notably, the two factors can also induce pauses. We further report that EfpL can sense the metabolic state of the cell via lysine acylation. Overall, our work characterizes the role of EfpL in ribosome rescue at proline-containing sequences, and provides evidence that co-occurrence of EF-P and EfpL is an evolutionary driver for higher bacterial growth rates.