2024-12-19 カリフォルニア大学サンディエゴ校(UCSD)
<関連情報>
- https://today.ucsd.edu/story/new-software-unlocks-secrets-of-cell-signaling
- https://www.nature.com/articles/s43588-024-00745-x
生体細胞における反応と輸送の空間モデリングアルゴリズム Spatial modeling algorithms for reactions and transport in biological cells
Emmet A. Francis,Justin G. Laughlin,Jørgen S. Dokken,Henrik N. T. Finsberg,Christopher T. Lee,Marie E. Rognes & Padmini Rangamani,
Nature Computational Science Published:19 December 2024
DOI:https://doi.org/10.1038/s43588-024-00745-x
A preprint version of the article is available at bioRxiv.
Abstract
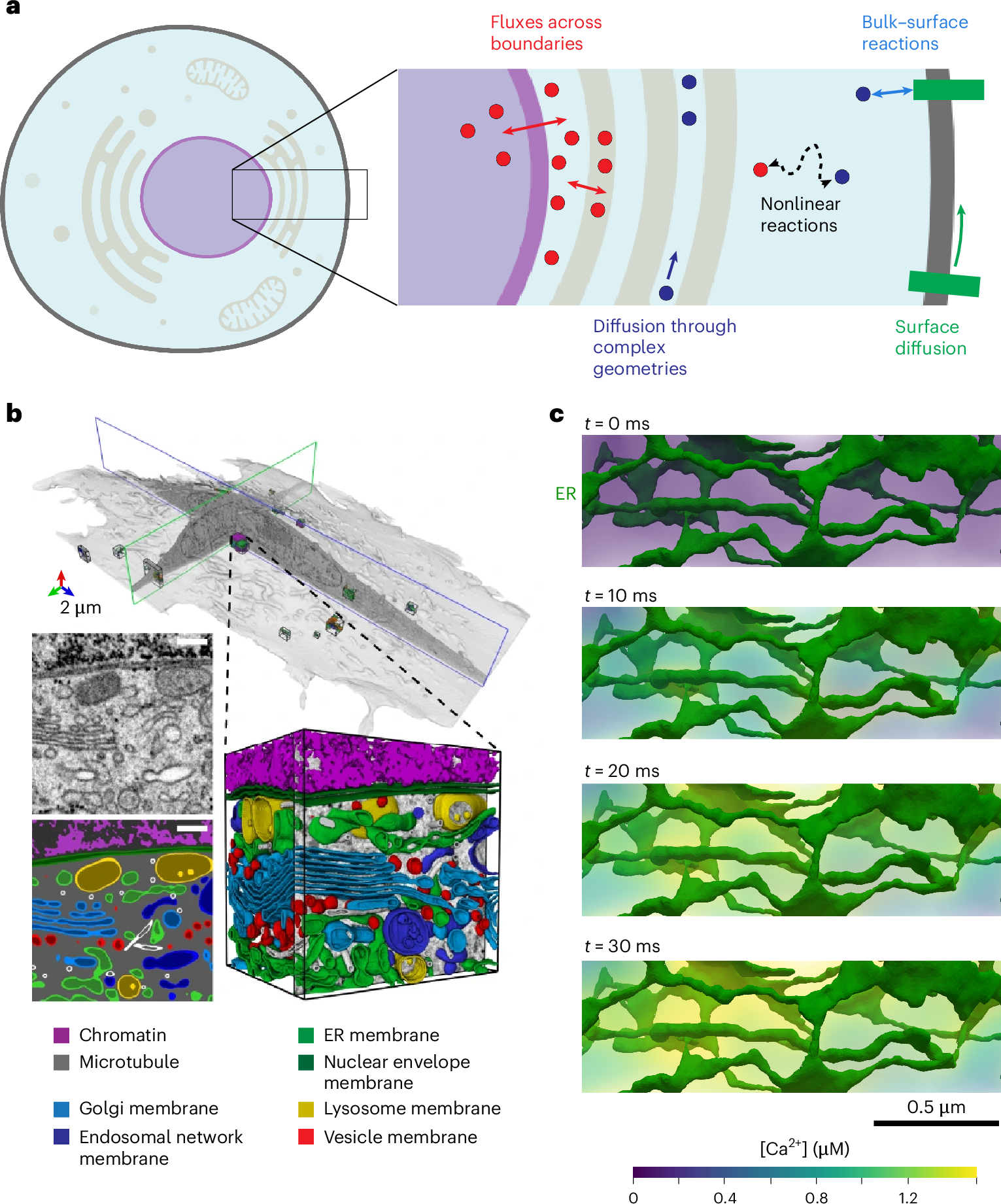
Biological cells rely on precise spatiotemporal coordination of biochemical reactions to control their functions. Such cell signaling networks have been a common focus for mathematical models, but they remain challenging to simulate, particularly in realistic cell geometries. Here we present Spatial Modeling Algorithms for Reactions and Transport (SMART), a software package that takes in high-level user specifications about cell signaling networks and then assembles and solves the associated mathematical systems. SMART uses state-of-the-art finite element analysis, via the FEniCS Project software, to efficiently and accurately resolve cell signaling events over discretized cellular and subcellular geometries. We demonstrate its application to several different biological systems, including yes-associated protein (YAP)/PDZ-binding motif (TAZ) mechanotransduction, calcium signaling in neurons and cardiomyocytes, and ATP generation in mitochondria. Throughout, we utilize experimentally derived realistic cellular geometries represented by well-conditioned tetrahedral meshes. These scenarios demonstrate the applicability, flexibility, accuracy and efficiency of SMART across a range of temporal and spatial scales.

