2025-01-08 コロンビア大学
<関連情報>
- https://www.cuimc.columbia.edu/news/new-ai-predicts-inner-workings-cells#
- https://www.nature.com/articles/s41586-024-08391-z
ヒト細胞型における転写の基礎モデル A foundation model of transcription across human cell types
Xi Fu,Shentong Mo,Alejandro Buendia,Anouchka P. Laurent,Anqi Shao,Maria del Mar Alvarez-Torres,Tianji Yu,Jimin Tan,Jiayu Su,Romella Sagatelian,Adolfo A. Ferrando,Alberto Ciccia,Yanyan Lan,David M. Owens,Teresa Palomero,Eric P. Xing & Raul Rabadan
Nature Published:08 January 2025
DOI:https://doi.org/10.1038/s41586-024-08391-z
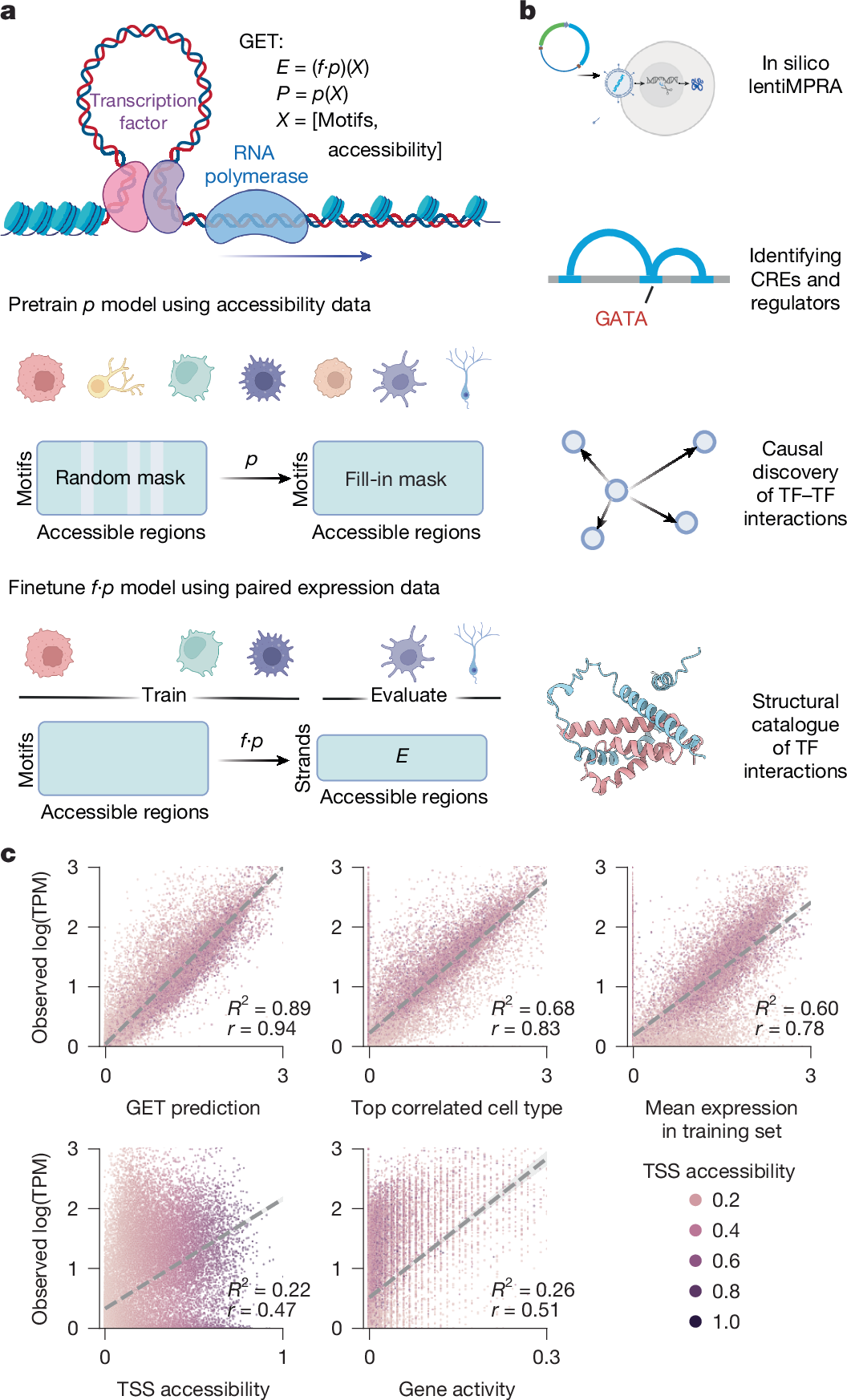
Abstract
Transcriptional regulation, which involves a complex interplay between regulatory sequences and proteins, directs all biological processes. Computational models of transcription lack generalizability to accurately extrapolate to unseen cell types and conditions. Here we introduce GET (general expression transformer), an interpretable foundation model designed to uncover regulatory grammars across 213 human fetal and adult cell types1,2. Relying exclusively on chromatin accessibility data and sequence information, GET achieves experimental-level accuracy in predicting gene expression even in previously unseen cell types3. GET also shows remarkable adaptability across new sequencing platforms and assays, enabling regulatory inference across a broad range of cell types and conditions, and uncovers universal and cell-type-specific transcription factor interaction networks. We evaluated its performance in prediction of regulatory activity, inference of regulatory elements and regulators, and identification of physical interactions between transcription factors and found that it outperforms current models4 in predicting lentivirus-based massively parallel reporter assay readout5,6. In fetal erythroblasts7, we identified distal (greater than 1 Mbp) regulatory regions that were missed by previous models, and, in B cells, we identified a lymphocyte-specific transcription factor–transcription factor interaction that explains the functional significance of a leukaemia risk predisposing germline mutation8,9,10. In sum, we provide a generalizable and accurate model for transcription together with catalogues of gene regulation and transcription factor interactions, all with cell type specificity.