2025-04-22 カリフォルニア工科大学 (Caltech)
<関連情報>
- https://www.caltech.edu/about/news/a-new-tool-to-detect-viruses-in-sequence-data
- https://www.nature.com/articles/s41587-025-02614-y
単一細胞の分解能でウイルス配列を検出し、宿主の遺伝子発現変化に関連する新規ウイルスを同定する Detection of viral sequences at single-cell resolution identifies novel viruses associated with host gene expression changes
Laura Luebbert,Delaney K. Sullivan,Maria Carilli,Kristján Eldjárn Hjörleifsson,Alexander Viloria Winnett,Tara Chari & Lior Pachter
Nature Biotechnology Published:22 April 2025
DOI:https://doi.org/10.1038/s41587-025-02614-y
Abstract
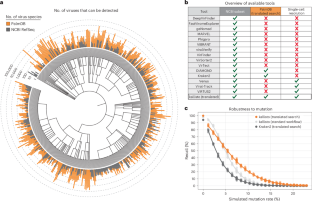
The increasing use of high-throughput sequencing methods in research, agriculture and healthcare provides an opportunity for the cost-effective surveillance of viral diversity and investigation of virus–disease correlation. However, existing methods for identifying viruses in sequencing data rely on and are limited to reference genomes or cannot retain single-cell resolution through cell barcode tracking. We introduce a method that accurately and rapidly detects viral sequences in bulk and single-cell transcriptomics data based on the highly conserved RdRP protein, enabling the detection of over 100,000 RNA virus species. The analysis of viral presence and host gene expression in parallel at single-cell resolution allows for the characterization of host viromes and the identification of viral tropism and host responses. We apply our method to peripheral blood mononuclear cell data from rhesus macaques with Ebola virus disease and describe previously unknown putative viruses. Moreover, we are able to accurately predict viral presence in individual cells based on macaque gene expression.