2025-05-09 イェール大学
<関連情報>
- https://www.yalecancercenter.org/news-article/study-reveals-key-insights-into-glioblastoma-recurrence-possible-therapy-targets/
- https://www.nature.com/articles/s41588-025-02167-5
- https://www.nature.com/articles/s41588-025-02168-4
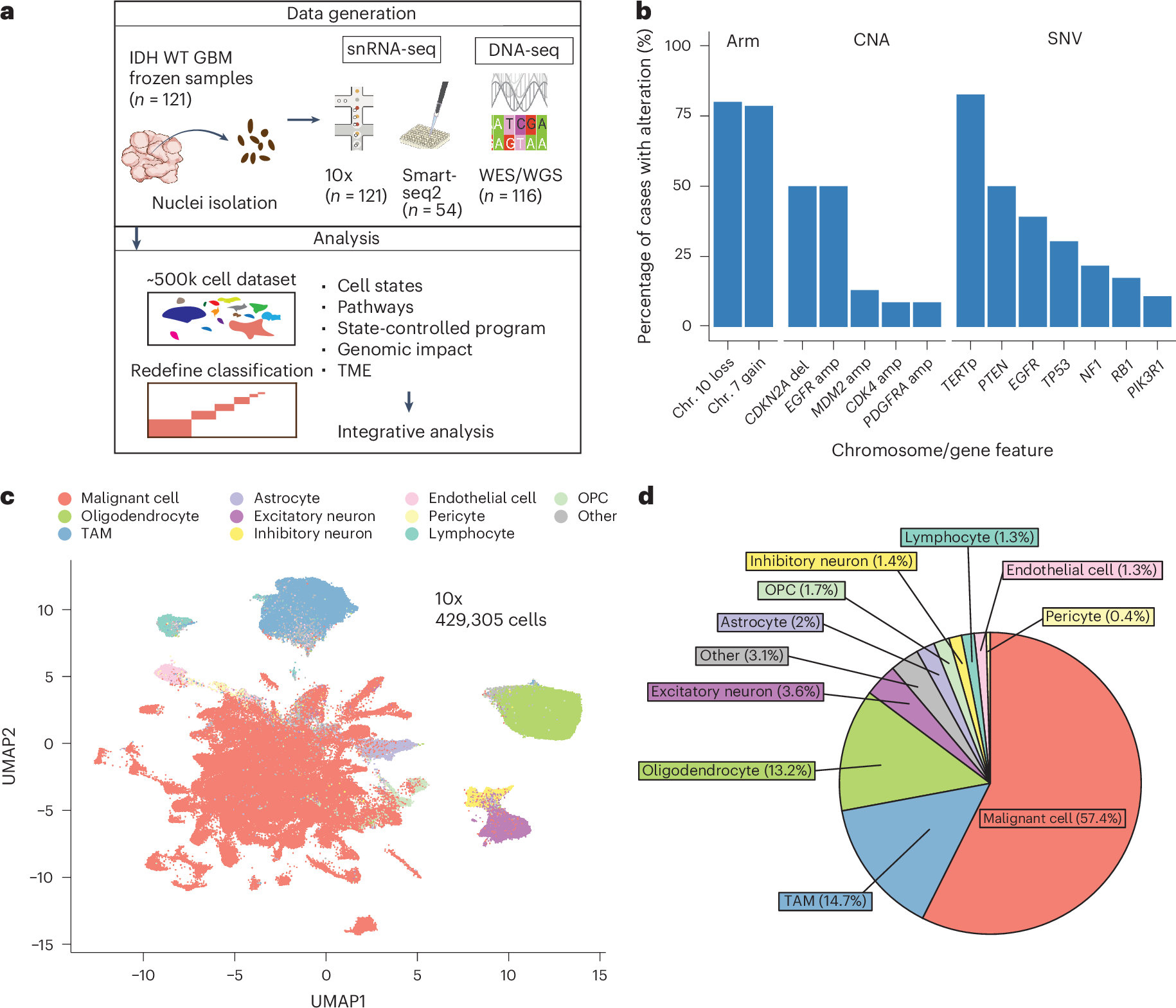
膠芽腫エコシステムの多層転写構造 The multilayered transcriptional architecture of glioblastoma ecosystems
Masashi Nomura,Avishay Spitzer,Kevin C. Johnson,Luciano Garofano,Djamel Nehar-belaid,Noam Galili Darnell,Alissa C. Greenwald,Lillian Bussema,Young Taek Oh,Frederick S. Varn,Fulvio D’Angelo,Simon Gritsch,Kevin J. Anderson,Simona Migliozzi,L. Nicolas Gonzalez Castro,Tamrin ChowdhFury,Nicolas Robine,Catherine Reeves,Jong Bae Park,Anuja Lipsa,Frank Hertel,Anna Golebiewska,Simone P. Niclou,Labeeba Nusrat,… Itay Tirosh
Nature Genetics Published:09 May 2025
DOI:https://doi.org/10.1038/s41588-025-02167-5
Abstract
In isocitrate dehydrogenase wildtype glioblastoma (GBM), cellular heterogeneity across and within tumors may drive therapeutic resistance. Here we analyzed 121 primary and recurrent GBM samples from 59 patients using single-nucleus RNA sequencing and bulk tumor DNA sequencing to characterize GBM transcriptional heterogeneity. First, GBMs can be classified by their broad cellular composition, encompassing malignant and nonmalignant cell types. Second, in each cell type we describe the diversity of cellular states and their pathway activation, particularly an expanded set of malignant cell states, including glial progenitor cell-like, neuronal-like and cilia-like. Third, the remaining variation between GBMs highlights three baseline gene expression programs. These three layers of heterogeneity are interrelated and partially associated with specific genetic aberrations, thereby defining three stereotypic GBM ecosystems. This work provides an unparalleled view of the multilayered transcriptional architecture of GBM. How this architecture evolves during disease progression is addressed in the companion manuscript by Spitzer et al.
統合的単一細胞ゲノミクスによる膠芽腫エコシステムの縦断的軌跡の解読 Deciphering the longitudinal trajectories of glioblastoma ecosystems by integrative single-cell genomics
Avishay Spitzer,Kevin C. Johnson,Masashi Nomura,Luciano Garofano,Djamel Nehar-belaid,Noam Galili Darnell,Alissa C. Greenwald,Lillian Bussema,Young Taek Oh,Frederick S. Varn,Fulvio D’Angelo,Simon Gritsch,Kevin J. Anderson,Simona Migliozzi,L. Nicolas Gonzalez Castro,Tamrin Chowdhury,Nicolas Robine,Catherine Reeves,Jong Bae Park,Anuja Lipsa,Frank Hertel,Anna Golebiewska,Simone P. Niclou,Labeeba Nusrat,… Mario L. Suvà
Nature Genetics Published:09 May 2025
DOI:https://doi.org/10.1038/s41588-025-02168-4
Abstract
The evolution of isocitrate dehydrogenase (IDH)-wildtype glioblastoma (GBM) after standard-of-care therapy remains poorly understood. Here we analyzed matched primary and recurrent GBMs from 59 patients using single-nucleus RNA sequencing and bulk DNA sequencing, assessing the longitudinal evolution of the GBM ecosystem across layers of cellular and molecular heterogeneity. The most consistent change was a lower malignant cell fraction at recurrence and a reciprocal increase in glial and neuronal cell types in the tumor microenvironment (TME). The predominant malignant cell state differed between most matched pairs, but no states were exclusive or highly enriched in either time point, nor was there a consistent longitudinal trajectory across the cohort. Nevertheless, specific trajectories were enriched in subsets of patients. Changes in malignant state abundances mirrored changes in TME composition and baseline profiles, reflecting the co-evolution of the GBM ecosystem. Our study provides a blueprint of GBM’s diverse longitudinal trajectories and highlights the treatment and TME modifiers that shape them.

