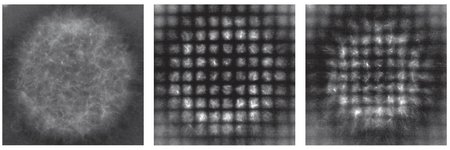
2025-06-09 ノースウェスタン大学
<関連情報>
- https://news.northwestern.edu/stories/2025/06/ai-identifies-key-gene-sets-that-cause-complex-disease/
- https://www.pnas.org/doi/10.1073/pnas.2415071122
複雑形質の原因遺伝子セットの生成的予測 Generative prediction of causal gene sets responsible for complex traits
Benjamin Kuznets-Speck, Buduka K. Ogonor, Thomas P. Wytock, and Adilson E. Motter
Proceedings of the National Academy of Sciences Published:June 12, 2025
DOI:https://doi.org/10.1073/pnas.2415071122

Significance
Researchers have long sought to bridge the gap between phenotypes and the genotypes that cause them. This gap remains open because current methods focus on associating phenotypes to a combinatorially explosive number of genotypic possibilities, resulting in a loss of statistical power. We overcome this limitation by employing transcriptomic data from complex, polygenic, human diseases combined with measured transcriptomic responses to gene perturbations in cell lines. The former data allow us to perform generative modeling and dimensional reduction to map transcriptome to phenotype, while the latter incorporate causal information regarding how gene regulation shapes phenotype. We predict sets of genes that explain the emergence of complex traits, which suggest possible multitarget disease treatments.
Abstract
The relationship between genotype and phenotype remains an outstanding question for organism-level traits because these traits are generally complex. The challenge arises from complex traits being determined by a combination of multiple genes (or loci), which leads to an explosion of possible genotype–phenotype mappings. The primary techniques to resolve these mappings are genome/transcriptome-wide association studies, which are limited by their lack of causal inference and statistical power. Here, we develop an approach that combines transcriptional data endowed with causal information and a generative machine learning model designed to strengthen statistical power. Our implementation of the approach—dubbed transcriptome-wide conditional variational autoencoder (TWAVE)—includes a variational autoencoder trained on human transcriptional data, which is incorporated into an optimization framework. Given a trait phenotype, TWAVE generates expression profiles, which we dimensionally reduce by identifying independently varying generalized pathways (eigengenes). We then conduct constrained optimization to find causal gene sets that are the gene perturbations whose measured transcriptomic responses best explain trait phenotype differences. By considering several complex traits, we show that the approach identifies causal genes that cannot be detected by the primary existing techniques. Moreover, the approach identifies complex diseases caused by distinct sets of genes, meaning that the disease is polygenic and exhibits distinct subtypes driven by different genotype–phenotype mappings. We suggest that the approach will enable the design of tailored experiments to identify multigenic targets to address complex diseases.