2025-07-29 ロックフェラー大学
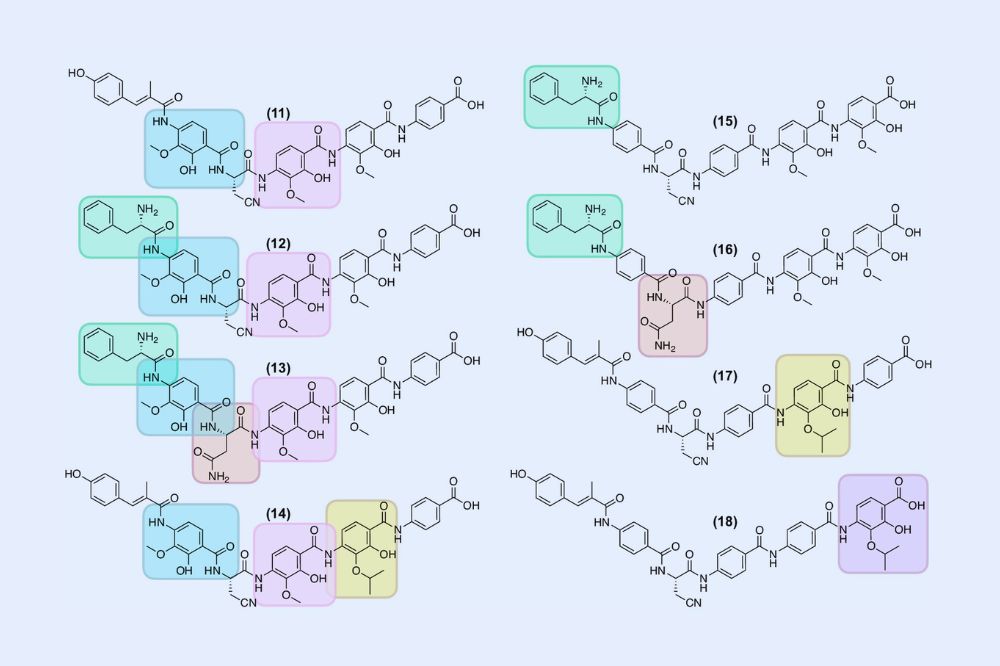
Scientists looked at variants of the antibiotic albicidin to identify structural differences (colored boxes) that may be associated with reduced suscepitibility to resistance. (Brady Lab)
<関連情報>
- https://www.rockefeller.edu/news/38082-new-tool-helps-predict-antibiotic-resistance/
- https://www.pnas.org/doi/10.1073/pnas.2504781122
環境耐性ゲノムに基づく耐性耐性抗生物質の開発 Environmental resistome–guided development of resistance-tolerant antibiotics
James Peek, Abir Bhattacharjee, Ján Burian, +4 , and Sean F. Brady
Proceedings of the National Academy of Sciences Published:May 19, 2025
DOI:https://doi.org/10.1073/pnas.2504781122
Significance
Many types of antibiotic resistance found in the clinic originated in the environment, yet the environment is largely overlooked as a potential source of future resistance threats for new antibiotics. We used metagenomic surveys of the environmental resistance reservoir (resistome) as an early warning system to anticipate future resistance problems for an antibiotic in development. This allowed us to proactively optimize the structure of the antibiotic to evade the forecasted resistance threats. Use of the environmental resistome to guide the optimization of the next generation of antibiotics would be a valuable addition to drug development programs to prolong the useful lifespans of the new compounds.
Abstract
Failure to anticipate new forms of antibiotic resistance has led to resistance developing rapidly to virtually all antibiotics that have entered clinical use. Many of the most problematic types of resistance originated in the environment, where ancient arms races between antibiotic-producing microbes and their competitors have created vast arsenals of antibiotics and resistance. Seizing on the knowledge that resistance in nature is frequently a harbinger of future clinical resistance, we propose introducing an additional step into the antibiotic development process that exploits the susceptibility of development candidates to environmental resistance as a metric for prioritizing lead compounds and as a roadmap for their structural optimization. Using the antibiotic albicidin as a model, we show how the environmental resistome can guide the development of more resistance-tolerant leads. We used metagenomic surveys to identify resistance vulnerabilities for albicidin and guide the synthesis of analogs that evade the resistance threats. We found that natural albicidin analogs (congeners) were especially enriched in structural features that escape resistance, which inspired our syntheses and provided compelling evidence for the evolution of families of antibiotics in response to resistance in nature. The coupling of metagenomics-based resistance surveillance with structural optimizations of new antibiotics is a broadly applicable approach that is easily integrated into antibiotic development programs to generate compounds that are more resilient in the face of resistance.