2025-10-20 コロンビア大学
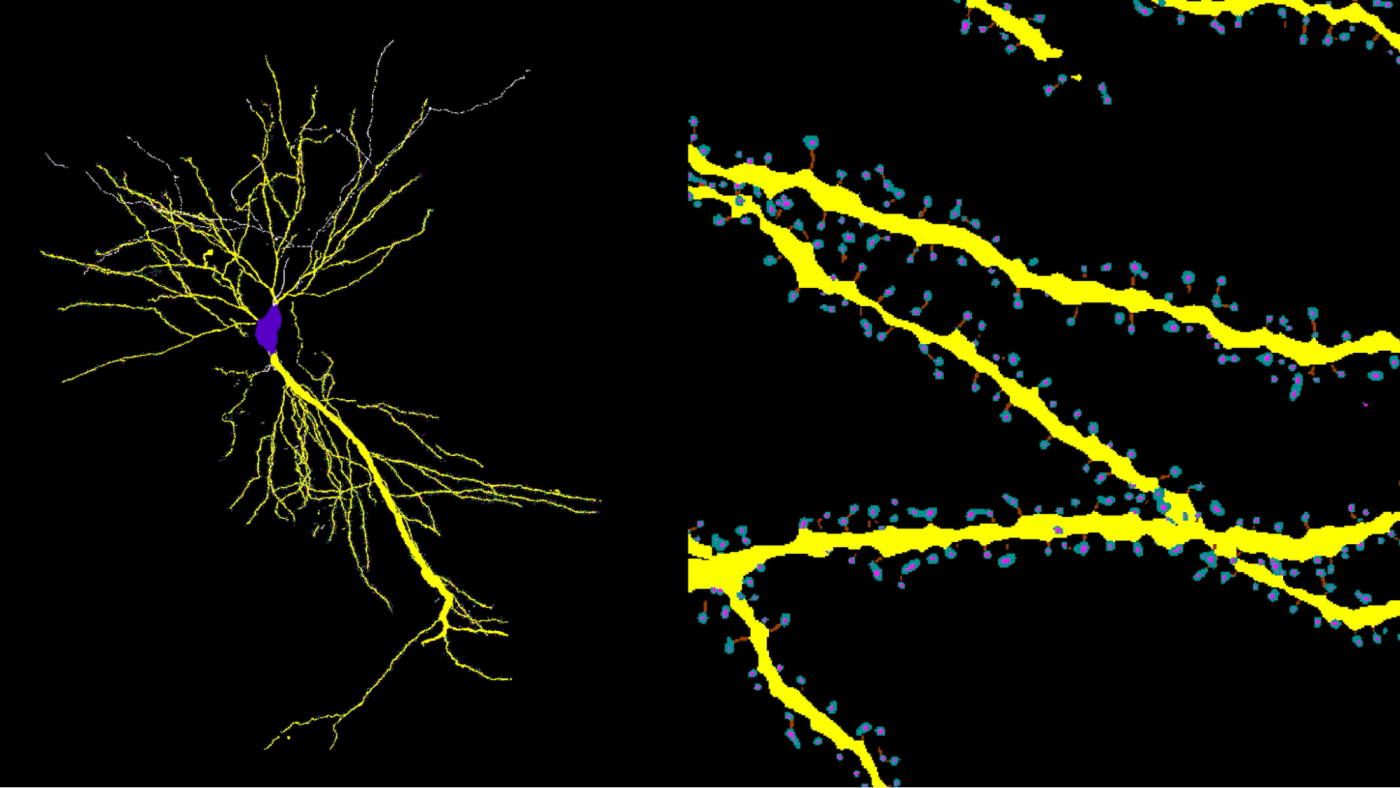
RESPAN maps thousands of excitatory synapses (spines) along the dendritic arbor (yellow) of a CA1 pyramidal neuron. (Credit: Kevin Gonzalez, Sergio Bernal-Garcia / Polleux lab / Zuckerman Institute and Luke Hammond / Ohio State University)
<関連情報>
- https://zuckermaninstitute.columbia.edu/ai-tool-reveals-complete-3d-architecture-brain-cells
- https://www.sciencedirect.com/science/article/pii/S2667237525002152
樹状突起棘の正確かつ自動化された復元、セグメンテーション、定量化のためのディープラーニングパイプライン A deep learning pipeline for accurate and automated restoration, segmentation, and quantification of dendritic spines
Sergio Bernal-Garcia, Alexa P. Schlotter, Daniela B. Pereira, Aleksandra J. Recupero, Franck Polleux, Luke A. Hammond
Cell Reports Methods Available online: 18 September 2025
DOI:https://doi.org/10.1016/j.crmeth.2025.101179
Highlights
- RESPAN is user friendly and allows accurate and unbiased dendritic spine analysis
- RESPAN integrates deep learning restoration, segmentation, and extensive quantification
- RESPAN analyzes data with heterogeneous image quality without parameter tuning
Motivation
Accurate and unbiased reconstructions of neuronal morphology and quantification of dendritic spines are widely used in neuroscience but remain a significant challenge for efficient large-scale analysis. Current methods rely heavily on manual annotation and parameter optimization between images, introducing bias and creating bottlenecks that limit large-scale studies. Additionally, existing automated tools often require complex workflows across multiple software platforms and lack integrated validation capabilities. We developed RESPAN to address these limitations by providing a comprehensive, automated pipeline that combines state-of-the-art deep learning approaches for image restoration and segmentation, model training, analysis, and validation within a single user-friendly graphical interface. This enables rapid, unbiased analysis of dendritic spine morphology across diverse imaging modalities while maintaining high accuracy and reproducibility.

