2026-02-11 パシフィック・ノースウェスト国立研究所(PNNL)
Understanding molecular modifications in red yeast could help scientists re-engineer the organism’s phenotype.(Image by Stephanie King | Pacific Northwest National Laboratory)
<関連情報>
- https://www.pnnl.gov/publications/cyanobacteria-rapidly-adapt-environmental-perturbations-through-structural-remodeling
- https://www.sciencedirect.com/science/article/pii/S1535947625005420
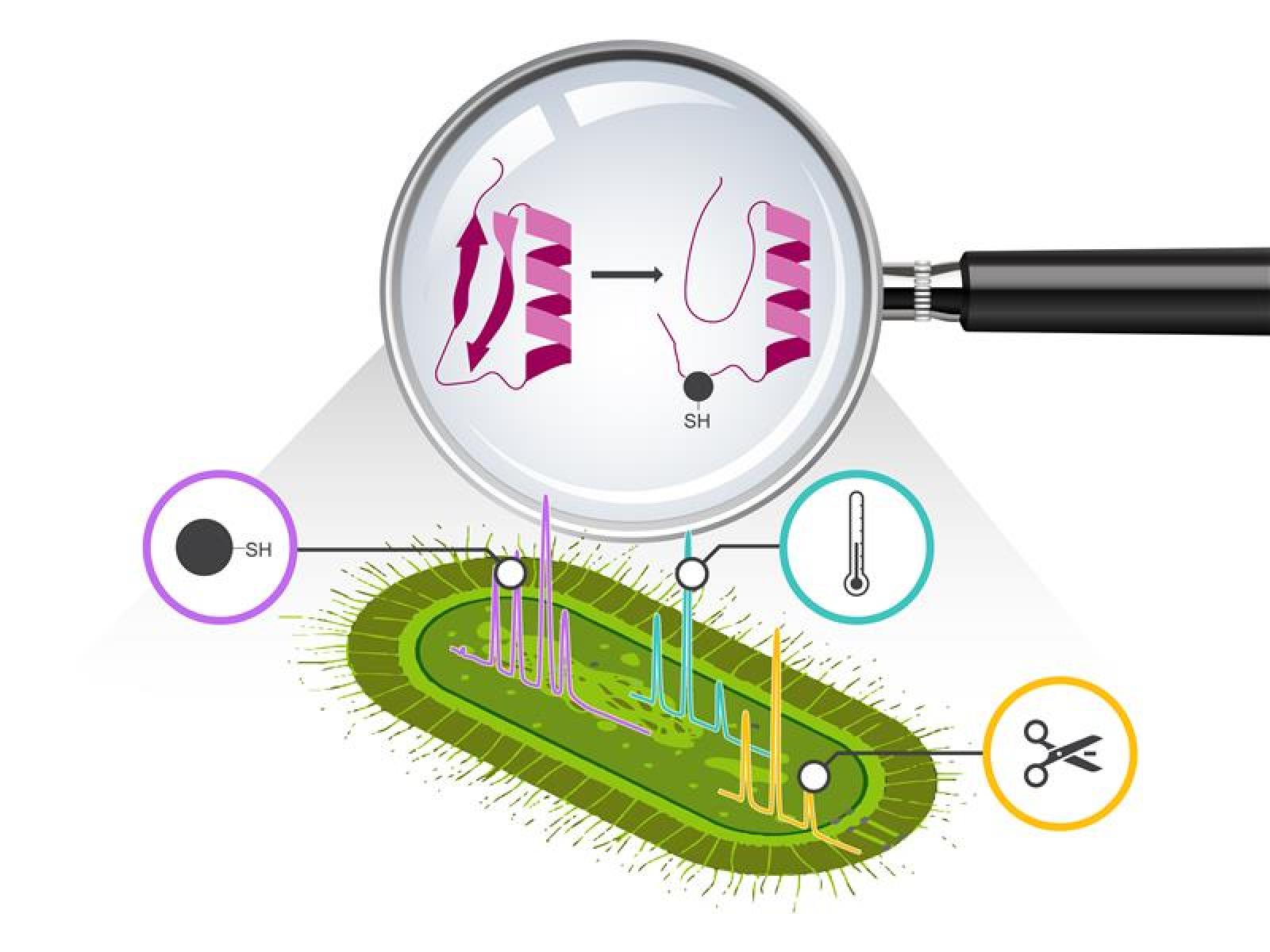
シアノバクテリアの環境変動への迅速な適応は、プロテオームの構造リモデリングによって達成される Rapid adaptation of cyanobacteria to environmental perturbations is achieved through structural remodeling of the proteome
Snigdha Sarkar, Elise M. Van Fossen, Xiaolu Li, Tong Zhang, Song Feng, Victoria Prozapas, Ivo Díaz Ludovico, Abdullah D. Shouaib, Chelsea M. Hutchinson-Bunch, Natalie C. Sadler, Isaac K. Attah, Wei-Jun Qian, Margaret S. Cheung, Pavlo Bohutskyi, John T. Melchior
Molecular & Cellular Proteomics Available online: 3 November 2025
DOI:https://doi.org/10.1016/j.mcpro.2025.101443
Highlights
- Structural proteomics reveal how cyanobacteria adapt to changing light.
- Light shifts trigger remodeling of phycobilisome and electron transport proteins.
- Data integration shows how photosynthesis regulates carbon metabolism pathways
Abstract
Dynamic environments require cyanobacteria to rapidly respond to fluctuating light conditions on timescales faster than transcription-translation processes allow, which is possible through immediate regulation of protein function via molecular and conformational adjustments. Traditional abundance-based proteomics cannot capture these rapid structural changes, creating a critical gap in understanding cellular adaptation mechanisms. We hypothesized that application of alternative structural proteomics approaches would enable identification of immediate and extensive structural remodeling across the cyanobacterial proteome triggered by environmental perturbations, potentially driving functional adaptations invisible to conventional abundance-based methods. We interrogated three complementary techniques—limited proteolysis mass spectrometry (LiP-MS), thermal proteome profiling (TPP-MS), and redox proteomics—for their capacity to unveil structural reorganization within the model cyanobacterium Synechococcus elongatus PCC 7942 during physiologically relevant light transitions. Within 30 minutes of increased light exposure, we detected structural changes in 753 proteins (LiP-MS), thermal stability shifts in 600 proteins (TPP-MS), and cysteine oxidation in 1,887 sites, while only 145 proteins changed in abundance. All three techniques consistently revealed coordinated remodeling of photosynthetic machinery, ribosomal complexes, and carbon metabolism, exemplified by cytochrome f stabilization modulating electron transport efficiency. Remarkably, <10% of proteins overlapped between methods, demonstrating that each technique captures distinct molecular dimensions of environmental adaptation. This structural proteomics framework demonstrates how alternative techniques can reveal hidden facets of proteome dynamics underlying cellular processes, offering new methodological approaches for understanding environmental responses and informing biotechnological applications.


