ピッツ大学研究者、計算モデルにより、一部のインフルエンザウイルスが重症化する理由を解明 Pitt Researchers Use Computational Modeling to Understand Why Some Flu Viruses Cause More Severe Infections
2022-07-21 ピッツバーグ大学
高病原性(病気を引き起こす)鳥インフルエンザであるH5N1と、低病原性の豚インフルエンザであるH1N1に感染したマウスからのデータを使いました。その後、工学的アプローチを用いて、ウイルスの複製と、インターフェロンや免疫細胞の活性レベルなど、マウスの感染に基づく主要な免疫反応をモデル化し、予測した。さまざまな生物学的反応を調べることで、研究者たちは、インターフェロンの産生速度が、マウスに観察される株特異的な免疫反応を引き起こすことを突き止めた。
<関連情報>
数理モデルによるインフルエンザ感染時のインターフェロン産生速度の違いによる株特異的な免疫ダイナミクスの発見 Mathematical Modeling Finds Disparate Interferon Production Rates Drive Strain-Specific Immunodynamics during Deadly Influenza Infection
Emily E. Ackerman,Jordan J. A. Weaver and Jason E. Shoemaker
Viruses Published: 27 April 2022
DOI:https://doi.org/10.3390/v14050906
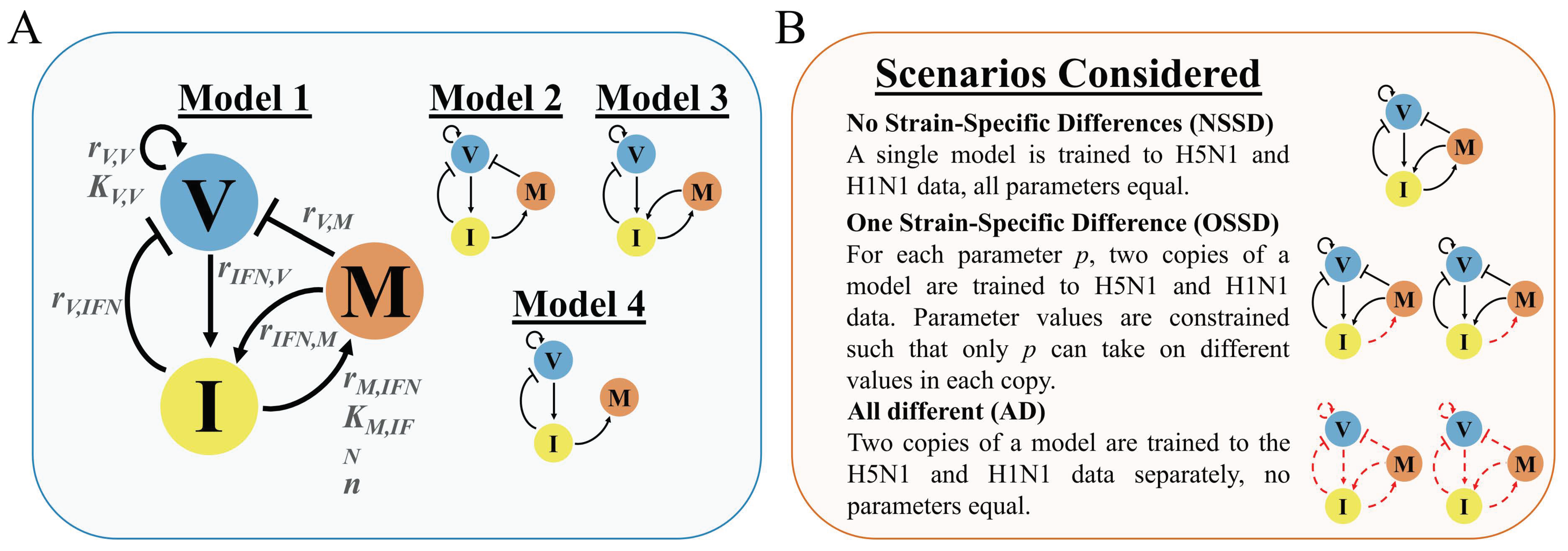
Abstract
The timing and magnitude of the immune response (i.e., the immunodynamics) associated with the early innate immune response to viral infection display distinct trends across influenza A virus subtypes in vivo. Evidence shows that the timing of the type-I interferon response and the overall magnitude of immune cell infiltration are both correlated with more severe outcomes. However, the mechanisms driving the distinct immunodynamics between infections of different virus strains (strain-specific immunodynamics) remain unclear. Here, computational modeling and strain-specific immunologic data are used to identify the immune interactions that differ in mice infected with low-pathogenic H1N1 or high-pathogenic H5N1 influenza viruses. Computational exploration of free parameters between strains suggests that the production rate of interferon is the major driver of strain-specific immune responses observed in vivo, and points towards the relationship between the viral load and lung epithelial interferon production as the main source of variance between infection outcomes. A greater understanding of the contributors to strain-specific immunodynamics can be utilized in future efforts aimed at treatment development to improve clinical outcomes of high-pathogenic viral strains.