20024-05-28 バッファロー大学(UB)
<関連情報>
- https://www.buffalo.edu/news/releases/2024/05/UB-research-RNA-editing-sites.html
- https://www.nature.com/articles/s42003-024-06239-w
APOBEC3を介したC-to-U RNA編集のヒト疾患への影響 The implications of APOBEC3-mediated C-to-U RNA editing for human disease
Melissa Van Norden,Zackary Falls,Sapan Mandloi,Brahm H. Segal,Bora E. Baysal,Ram Samudrala & Peter L. Elkin
Communications Biology Published:04 May 2024
DOI:https://doi.org/10.1038/s42003-024-06239-w
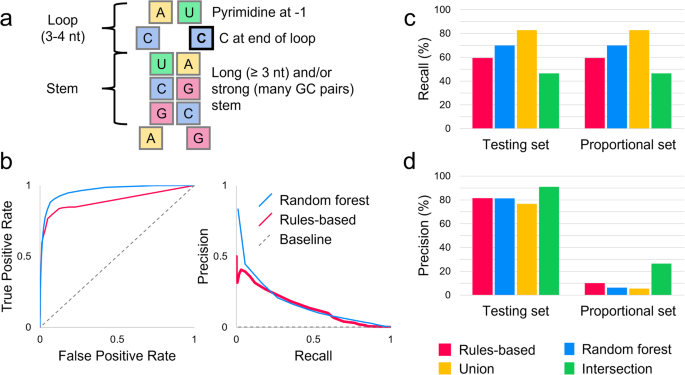
Abstract
Intra-organism biodiversity is thought to arise from epigenetic modification of constituent genes and post-translational modifications of translated proteins. Here, we show that post-transcriptional modifications, like RNA editing, may also contribute. RNA editing enzymes APOBEC3A and APOBEC3G catalyze the deamination of cytosine to uracil. RNAsee (RNA site editing evaluation) is a computational tool developed to predict the cytosines edited by these enzymes. We find that 4.5% of non-synonymous DNA single nucleotide polymorphisms that result in cytosine to uracil changes in RNA are probable sites for APOBEC3A/G RNA editing; the variant proteins created by such polymorphisms may also result from transient RNA editing. These polymorphisms are associated with over 20% of Medical Subject Headings across ten categories of disease, including nutritional and metabolic, neoplastic, cardiovascular, and nervous system diseases. Because RNA editing is transient and not organism-wide, future work is necessary to confirm the extent and effects of such editing in humans.