2025-03-18 産業技術総合研究所
<関連情報>
- https://www.aist.go.jp/aist_j/press_release/pr2025/pr20250318/pr20250318.html
- https://academic.oup.com/ismecommun/article/5/1/ycaf028/8008921
rRNA遺伝子アンプリコンシーケンスによるマイクロバイオームのクロスドメイン絶対定量化のための合成DNAスパイクイン標準物質 Synthetic DNA spike-in standards for cross-domain absolute quantification of microbiomes by rRNA gene amplicon sequencing
Dieter M Tourlousse, Yuji Sekiguchi
ISME Communications Published:17 March 2025
DOI:https://doi.org/10.1093/ismeco/ycaf028
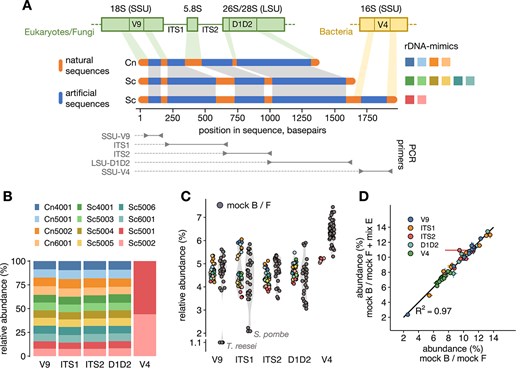
Abstract
Microbiome studies using high-throughput sequencing are increasingly incorporating absolute quantitative approaches to overcome the inherent limitations of relative abundances. In this study, we have designed and experimentally validated a set of 12 unique synthetic rRNA operons, which we refer to as rDNA-mimics, to serve as spike-in standards for quantitative profiling of fungal/eukaryotic and bacterial microbiomes. The rDNA-mimics consist of conserved sequence regions from natural rRNA genes to act as binding sites for common universal PCR primers, and bioinformatically designed variable regions that allow their robust identification in any microbiome sample. All constructs cover multiple rRNA operon regions commonly targeted in fungal/eukaryotic microbiome studies (SSU-V9, ITS1, ITS2, and LSU-D1D2) and two of them also include an artificial segment of the bacterial 16S rRNA gene (SSU-V4) for cross-domain application. We validated the quantitative performance of the rDNA-mimics using defined mock communities and representative environmental samples. In particular, we show that rDNA-mimics added to extracted DNA or directly to the samples prior to DNA extraction precisely reflects the total amount of fungal and/or bacterial rRNA genes in the samples. We demonstrate that this allows accurate estimation of differences in microbial loads between samples, thereby confirming that the rDNA-mimics are suitable for absolute quantitative analyses of differential microbial abundances.