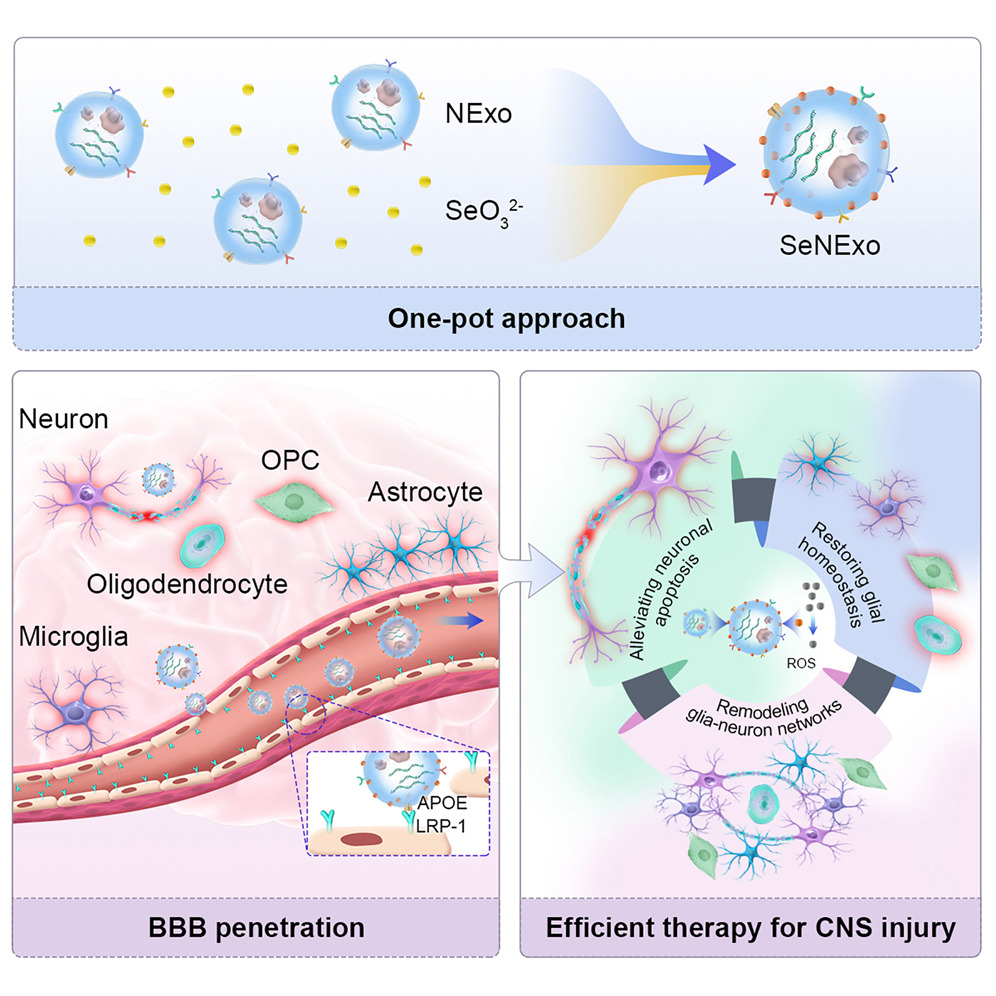
2025-08-22 中国科学院(CAS)
Web要約 の発言:
<関連情報>
- https://english.cas.cn/newsroom/research_news/life/202508/t20250826_1051372.shtml
- https://onlinelibrary.wiley.com/doi/10.1002/anie.202511839
創薬のためのDNAエンコードライブラリーにおける合理的な設計戦略 Rational Design Strategies in DNA-Encoded Libraries for Drug Discovery
Xudong Wang, Linjie Li, Xuanjing Shen, Prof. Xiaojie Lu
Angewandte Chemie International Edition Published: 25 July 2025
DOI:https://doi.org/10.1002/anie.202511839
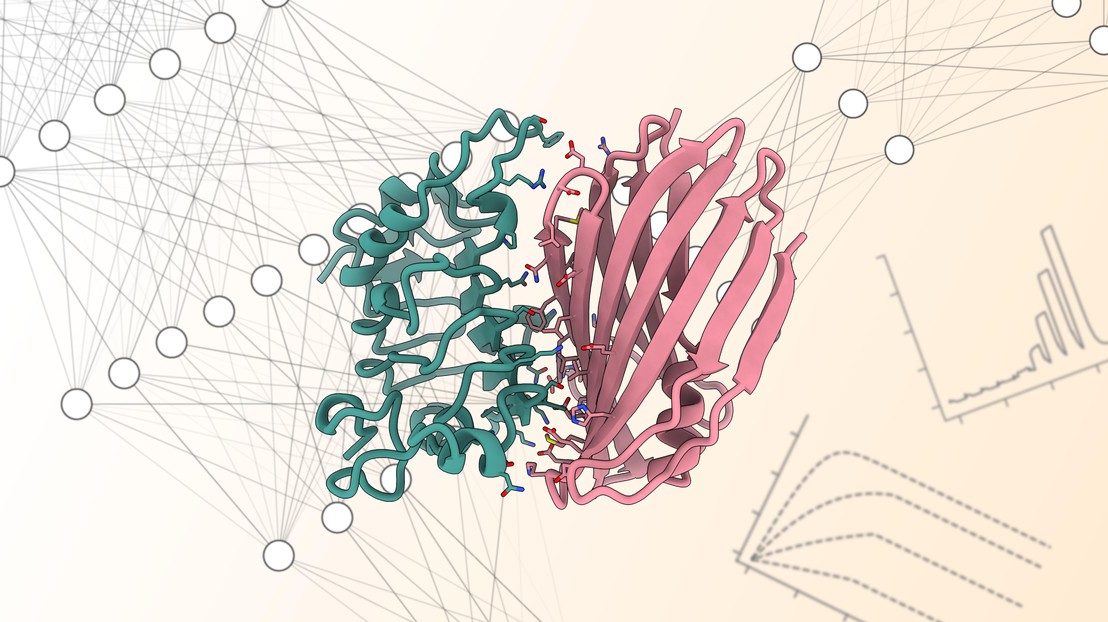
Graphical Abstract
This review highlights recent advances in rational DNA-encoded library (DEL) design, focusing on fragment-based DELs, covalent DELs, and focused DELs. These innovations shift DELs from empirical screening to a more strategic, hypothesis-driven approach, enhancing hit quality and drug discovery efficiency.
Abstract
DNA-encoded libraries (DELs) have emerged as a powerful and cost-effective platform for high-throughput screening, enabling the rapid identification of small-molecule ligands against a wide range of biological targets. However, traditional DEL approaches often rely on empirical and broad-based library construction, which can lead to low hit rates, off-target interactions, and limited chemical diversity around pharmaceutically relevant motifs. Recent technological advances have sought to address these limitations, shifting DELs from a largely blind screening tool to a more rational and precision-oriented strategy. In this review, we systematically examine the evolution of DEL methodologies, with a particular focus on innovations in library design that enhance hit quality and screening efficiency. Specifically, we highlight the emergence of fragment-based DEL strategies for exploring chemical space with minimal structures, the incorporation of covalent warheads to enable irreversible binding to specific residues, and the development of focused DELs tailored to particular protein families or binding motifs. Together, these advances mark a shift from blind, empirical screening toward a more strategic and hypothesis-driven application of DEL technology.