2025-09-25 東京科学大学
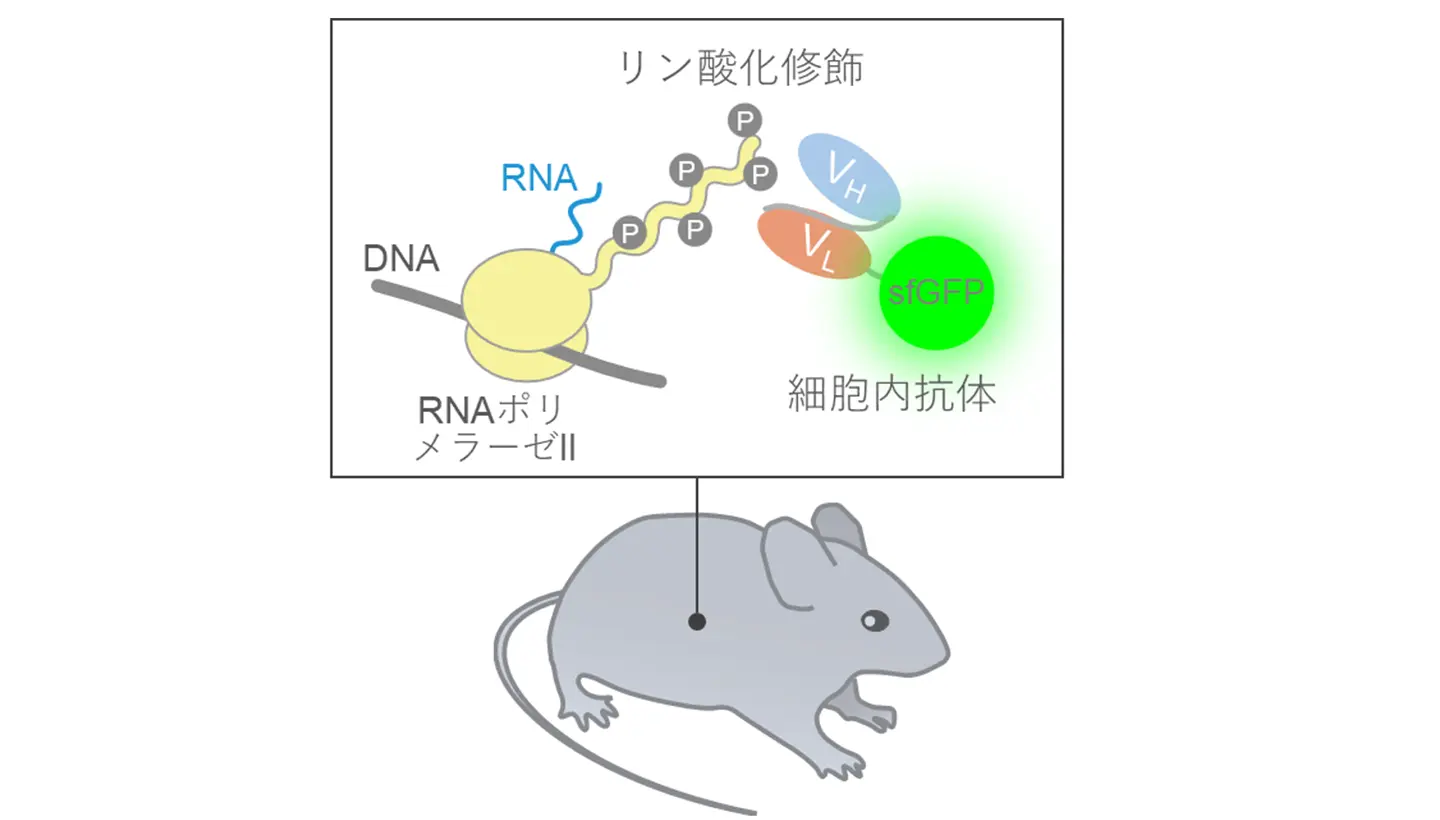
図1. RNAポリメラーゼIIによる転写の場所を可視化できるマウス
<関連情報>
- https://www.isct.ac.jp/ja/news/fadursnq0u44
- https://www.isct.ac.jp/plugins/cms/component_download_file.php?type=2&pageId=&contentsId=1&contentsDataId=2285&prevId=&key=0c06d6236d13da58b9a801e00a863980.pdf
- https://www.sciencedirect.com/science/article/abs/pii/S0022283625004619
マウス組織における転写伸長フォーカスの構成とダイナミクス Organization and Dynamics of Transcription Elongation Foci in Mouse Tissues
Chihiro Matsuda, Akane Ichiki, Yuko Sato, Yukino Kudo, Mika Saotome, Chihiro Takayama, Khoa Minh Le, Satoshi Uchino, Ryota Higuchi, Kazuhiko Kawata, Kosuke Tomimatsu, Manabu Ozawa, Masahito Ikawa, Yasuyuki Ohkawa, Yoshihiro Baba, Hiroshi Kimura
Journal of Molecular Biology Available online: 13 August 2025
DOI:https://doi.org/10.1016/j.jmb.2025.169395
Highlights
- RNA polymerase II elongation sites are visualized in live mouse tissues using a Ser2ph-specific intracellular antibody probe.
- The number of transcription elongation foci varies across immune cell types.
- Transcription elongation foci are less mobile in differentiated cells compared to proliferating cells.
Abstract
RNA polymerase II (RNAP2) transcribes most genes in eukaryotic nuclei. During the transition from transcription initiation to productive elongation, and throughout the elongation phase, RNAP2 becomes phosphorylated at the Ser2 residue within the heptapeptide repeats of the carboxyl-terminal domain of its largest subunit. Antibodies specific to RNAP2 Ser2 phosphorylation (Ser2ph) have enabled visualization of active transcription sites in fixed cells and tissues. Here, we report the generation and characterization of knock-in mice ubiquitously expressing a fluorescent protein-tagged, modification-specific intracellular antibody (mintbody) targeting RNAP2 Ser2ph. Using these mice, we successfully visualized transcription elongation foci in mouse tissues and characterized their distribution and dynamics across diverse cell types. RNAP2 Ser2ph-mintbody formed hundreds to thousands of nuclear foci, which were excluded from heterochromatin and transcriptionally repressed domains, such as the XY body in pachytene spermatocytes. Quantitative analysis revealed tissue- and cell type-specific variation in both the number and mobility of transcription elongation foci. The mobility of transcription foci was more restricted in differentiated cells compared to differentiating and proliferating cells, likely reflecting a reduced number of actively transcribed genes and more limited open chromatin regions upon differentiation. These findings suggest that the spatial organization and dynamics of transcription elongation are closely associated with cell identity and differentiation status. The RNAP2 Ser2ph-mintbody knock-in mice provide a valuable tool for future studies of transcription organization and dynamics at the tissue level.