2025-12-15 三重大学
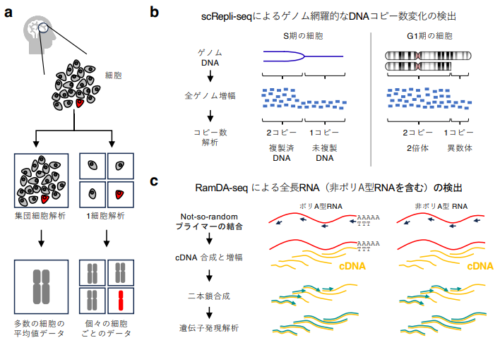
図1. 1細胞解析技術:scRepli-seqとRamDA-seqの概要
<関連情報>
- https://www.mie-u.ac.jp/news/topics/2025/12/post-3903.html
- https://www.mie-u.ac.jp/news/topics/09155a1e81c89f45c8a479fe8b028631.pdf
- https://www.nature.com/articles/s41467-025-64688-1
scRepli-RamDA-seq: S期における遺伝子発現動態の解析を可能にするマルチオミクス技術 scRepli-RamDA-seq: a multi-omics technology enabling the analysis of gene expression dynamics during S-phase
Rawin Poonperm,Taiki Yoneda,Taito Imada,Saori Takahashi,Takako Ichinose,Hisashi Miura,Tetsutaro Hayashi,Mariko Kuse,Mika Yoshimura,Koji Nagao,Chikashi Obuse,Itoshi Nikaido,Ichiro Hiratani & Shin-ichiro Takebayashi
Nature Communications Published:15 December 2025
DOI:https://doi.org/10.1038/s41467-025-64688-1
Abstract
Single-cell sequencing has advanced our understanding of cell-type diversity and heterogeneity. However, existing single-cell multi-omics methods lack the ability to monitor gene expression dynamics during S-phase progression. Here, we introduce single-cell (sc)Repli-RamDA-seq (scRR-seq), a multi-omics method that enables high-resolution DNA replication profiling and full-length total RNA sequencing from the same single cell in a haplotype-specific manner. scRR-seq generates DNA replication and RNA sequencing data comparable to individually obtained scRepli-seq and scRamDA-seq data, respectively. Unlike other scDNA/RNA-seq methods, scRR-seq allows one to tell the S-phase stage of a given cell based on the percentage of the replicated genome derived from scRepli-seq data. This facilitates analysis of gene expression dynamics during S-phase progression, enabling the identification of S-phase progression markers. scRR-seq also detects copy-number variation in non-S-phase cells and outperforms other scDNA/RNA-seq methods in various measures. Taken together, scRR-seq is a robust single-cell multi-omics method with promising potential for comprehensive genome/transcriptome analysis.


