2022-03-23 スウェーデン王・王立工科大学(KTH)
・TMEは、がんの発生に重要なさまざまな細胞で構成されています。これらの細胞の相互作用とアイデンティティを理解することは、腫瘍形成のメカニズムを発見するために不可欠である。
・TMEを研究するためには、細胞プロファイルに関する広範な情報とその空間的位置の両方を提供し、それらが互いにどのように相互作用するかを理解するための方法が必要です。
<関連情報>
- https://www.kth.se/en/om/mot/kalender/spatial-deconvolution-of-gene-expression-in-cancer-1.1145894?date=2022-03-25&orgdate=2022-03-23&length=1&orglength=0
- http://kth.diva-portal.org/smash/record.jsf?pid=diva2%3A1639909&dswid=4321
- http://kth.diva-portal.org/smash/get/diva2:1639909/FULLTEXT01.pdf
癌の空間的遺伝子発現のデコンボリューション Deconvolution of Spatial Gene Expression in Cancer
Abstract:
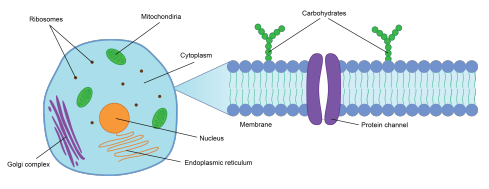
Cancer is the second leading cause of death in the world, claiming nearly 10 million lives in 2020 alone. One of the main issues in anti-cancer treatment is the heterogeneity of the tumor microenvironment (TME). The TME consists of different cells that are critical for cancer development. Understanding the interactions and identity of these cells is vital to discovering the mechanisms for tumorigenesis. To fundamentally understand the development and mechanisms of the disease will help us in designing novel treatments moving forward. To study the TME, we need methods that both provide extensive information about the cellular profiles and their spatial location, in order to understand how they interact with each other. Single-cell RNA-seq (scRNA-seq) has provided extensive insights into the cellular composition of tumors. However, it requires dissociation of the cells and thus does not retain spatial information. There are several methods to study spatially resolved gene expression in tissues, but one that allows for untargeted and whole-transcriptome wide analysis is the in situ capturing method, Spatial transcriptomics (ST). Although this method allows us to know the location of the gene expression, the resolution is too low for single-cell analysis. With an initial capturing area of 100 μm, 3-30 cells are captured in each spot resulting in a mixture of cells giving rise to the gene expression. At this resolution, it is challenging to confidentially profile the cells, thus making it difficult to explore the cellular interactions fully. To fundamentally explore the TME, improvements need to be made.
In Paper I, we aimed to bridge the gap between ST and scRNA-seq by designing a new array with a capturing area of 2 μm. This new design increased the number of capture areas from 1007 to over 1.4 million and with over a 4000-fold improved resolution. We managed to get spatially resolved gene expression from mouse olfactory bulb (MOB) and breast tumor tissue at a sub-cellular resolution with this new design. Despite a low capture efficiency of around 1.3% per bead, we were able to identify differently expressed (DE) signatures specific to morphological layers, profile specific cell types and explore sub-cellular features. Paper II focuses on the information obtained from the widely available histological images. By integrating the spatial gene expression data from 23 different breast cancer patients with their morphological images via deep learning, we could predict gene expression on different samples solely from their histological images. This was further validated on external samples to ensure that it was applicable to other clinical data. In Paper III, we explored the biology of HER2-positive breast tumors by combining scRNA-seq with ST data from eight different HER2-positive patients. With this combinatorial approach, we studied the interactions of tumor-associated cell types and found tertiary lymphoid (TL)-like structures which have been shown to hold certain predictive power in treatment outcome. From this, we constructed a predictive model that could infer the presence of these TL-like structures across different tissue types and technical platforms. This was validated on external samples from breast cancer, rheumatoid arthritis and melanoma. Lastly, in Paper IV, we sought to improve upon the reproducibility and robustness of the method by automating the 10x Visium protocol on a robotic platform. To benchmark the protocol, we compared identical samples prepared both manually and with the automated approach and achieved high correlation scores of 0.995 and 0.990. By adapting the protocol on a Bravo Liquid Handling Platform, we were able to increase the throughput and robustness of the method and reduce hands-on time by over 80%.