2023-04-17 デューク大学(Duke)
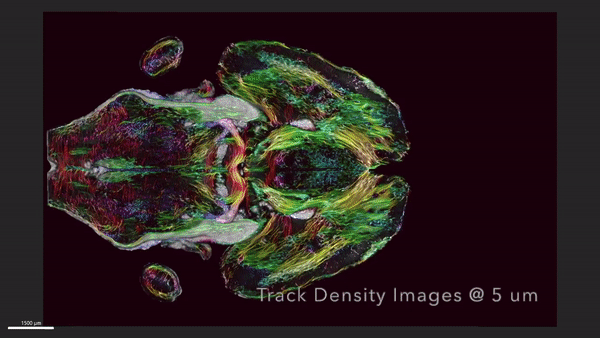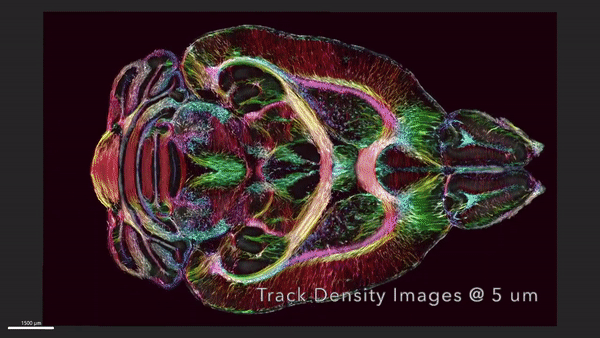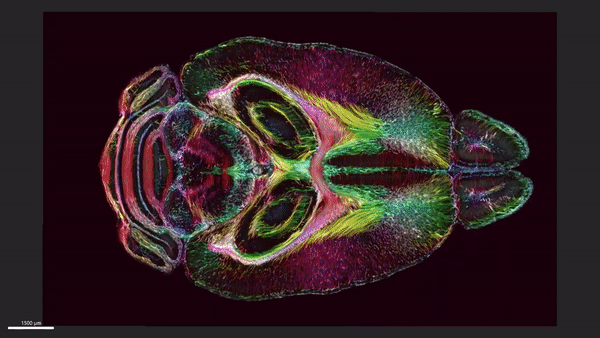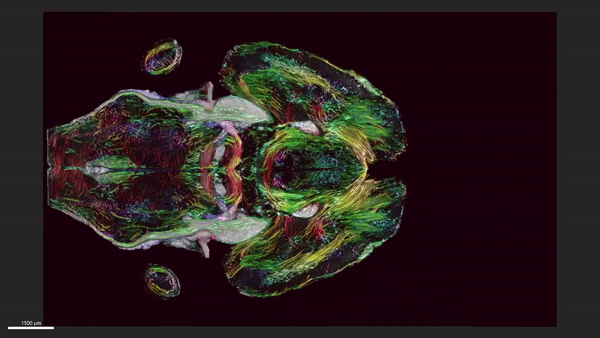
This video shows horizontal ‘slices’ of the circuitry data moving up and down across the brain.
<関連情報>
- https://today.duke.edu/2023/04/brain-images-just-got-64-million-times-sharper
- https://www.pnas.org/doi/10.1073/pnas.2218617120
マウス全脳の磁気共鳴と光シート顕微鏡法の融合 Merged magnetic resonance and light sheet microscopy of the whole mouse brain
G. Allan Johnson, Yuqi Tian, David G. Ashbrook, Gary P. Cofer, James J. Cook, James C. Gee, Adam Hall, Kathryn Hornburg, Yi Qi, Fang-Cheng Yeh, Nian Wang, Leonard E. White and Robert W. Williams
Proceedings of the National Academy of Sciences Published:April 17, 2023
DOI:https://doi.org/10.1073/pnas.2218617120
Significance
We demonstrate the highest-resolution MR images ever obtained of the mouse brain. The diffusion tensor images (DTI) @ 15 μm spatial resolution are 1,000 times the resolution of most preclinical rodent DTI/MRI. Superresolution track density images are 27,000 times that of typical preclinical DTI/MRI. High angular resolution yielded the most detailed MR connectivity maps ever generated. High-performance computing pipelines merged the DTI with light sheet microscopy of the same specimen, providing a comprehensive picture of cells and circuits. The methods have been used to demonstrate how strain differences result in differential changes in connectivity with age. We believe the methods will have broad applicability in the study of neurodegenerative diseases.
Abstract
We have developed workflows to align 3D magnetic resonance histology (MRH) of the mouse brain with light sheet microscopy (LSM) and 3D delineations of the same specimen. We start with MRH of the brain in the skull with gradient echo and diffusion tensor imaging (DTI) at 15 μm isotropic resolution which is ~ 1,000 times higher than that of most preclinical MRI. Connectomes are generated with superresolution tract density images of ~5 μm. Brains are cleared, stained for selected proteins, and imaged by LSM at 1.8 μm/pixel. LSM data are registered into the reference MRH space with labels derived from the ABA common coordinate framework. The result is a high-dimensional integrated volume with registration (HiDiver) with alignment precision better than 50 µm. Throughput is sufficiently high that HiDiver is being used in quantitative studies of the impact of gene variants and aging on mouse brain cytoarchitecture and connectomics.


