2023-09-14 ブラウン大学
◆ブラウン大学の研究チームは、トポロジーと呼ばれる数学の分野を活用し、胚における細胞の形状や空間パターンをプロファイル化する機械学習アルゴリズムを開発し、異なる細胞がどのように組み合わさるかを研究しています。
◆この研究は、細胞が異なる部位に配置されるメカニズムを探る新たな方向への道を開いています。
<関連情報>
- https://www.brown.edu/news/2023-09-14/tissue-architecture
- https://www.nature.com/articles/s41540-023-00302-8
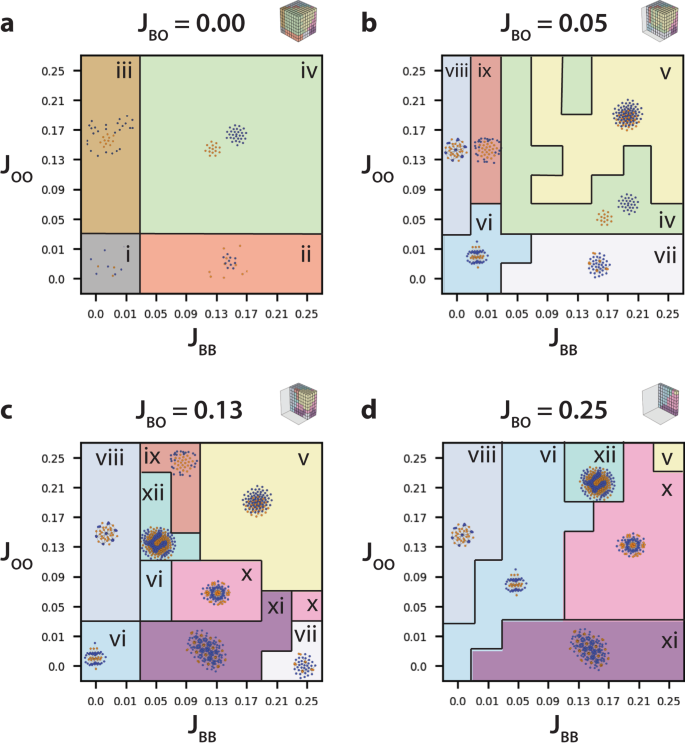
異種細胞集団における空間的パターン形成のトポロジーデータ解析:細胞間接着を変化させたクラスタリングとソーティング Topological data analysis of spatial patterning in heterogeneous cell populations: clustering and sorting with varying cell-cell adhesion
Dhananjay Bhaskar,William Y. Zhang,Alexandria Volkening,Björn Sandstede & Ian Y. Wong
npj Systems Biology and Applications Published:14 September 2023
DOI:https://doi.org/10.1038/s41540-023-00302-8
Abstract
Different cell types aggregate and sort into hierarchical architectures during the formation of animal tissues. The resulting spatial organization depends (in part) on the strength of adhesion of one cell type to itself relative to other cell types. However, automated and unsupervised classification of these multicellular spatial patterns remains challenging, particularly given their structural diversity and biological variability. Recent developments based on topological data analysis are intriguing to reveal similarities in tissue architecture, but these methods remain computationally expensive. In this article, we show that multicellular patterns organized from two interacting cell types can be efficiently represented through persistence images. Our optimized combination of dimensionality reduction via autoencoders, combined with hierarchical clustering, achieved high classification accuracy for simulations with constant cell numbers. We further demonstrate that persistence images can be normalized to improve classification for simulations with varying cell numbers due to proliferation. Finally, we systematically consider the importance of incorporating different topological features as well as information about each cell type to improve classification accuracy. We envision that topological machine learning based on persistence images will enable versatile and robust classification of complex tissue architectures that occur in development and disease.