2024-03-04 ジョージア工科大学
<関連情報>
- https://research.gatech.edu/whos-who-bacteria-reliable-way-define-species-and-strains
- https://www.nature.com/articles/s41467-023-44622-z
自然細菌集団を構成する菌株数の推定に向けて Towards estimating the number of strains that make up a natural bacterial population
Tomeu Viver,Roth E. Conrad,Luis M. Rodriguez-R,Ana S. Ramírez,Stephanus N. Venter,Jairo Rocha-Cárdenas,Mercè Llabrés,Rudolf Amann,Konstantinos T. Konstantinidis & Ramon Rossello-Mora
Nature Communications Published:16 January 2024
DOI:https://doi.org/10.1038/s41467-023-44622-z
Abstract
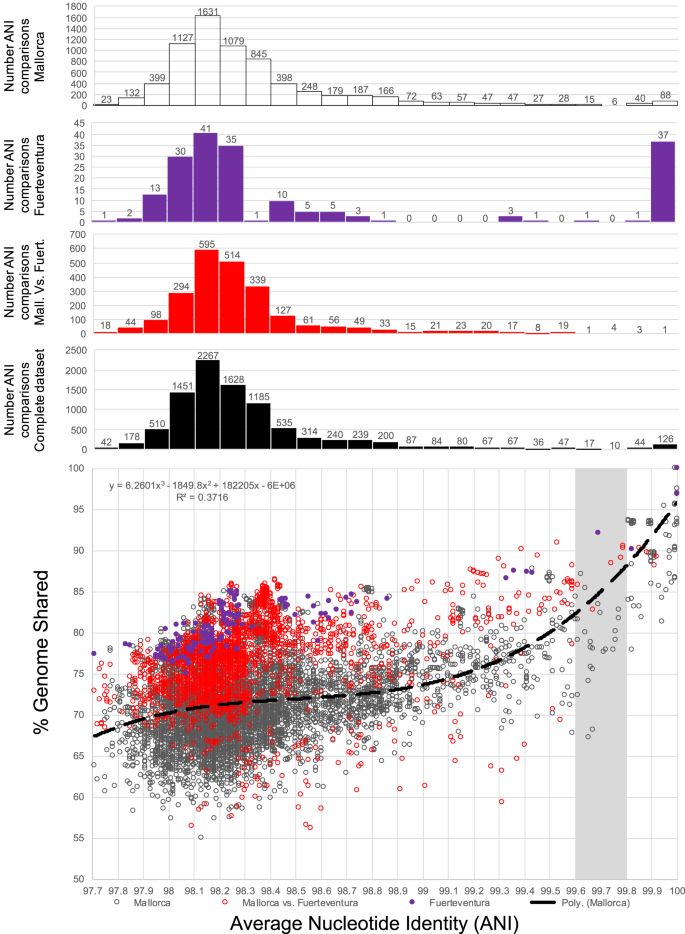
What a strain is and how many strains make up a natural bacterial population remain elusive concepts despite their apparent importance for assessing the role of intra-population diversity in disease emergence or response to environmental perturbations. To advance these concepts, we sequenced 138 randomly selected Salinibacter ruber isolates from two solar salterns and assessed these genomes against companion short-read metagenomes from the same samples. The distribution of genome-aggregate average nucleotide identity (ANI) values among these isolates revealed a bimodal distribution, with four-fold lower occurrence of values between 99.2% and 99.8% relative to ANI >99.8% or <99.2%, revealing a natural “gap” in the sequence space within species. Accordingly, we used this ANI gap to define genomovars and a higher ANI value of >99.99% and shared gene-content >99.0% to define strains. Using these thresholds and extrapolating from how many metagenomic reads each genomovar uniquely recruited, we estimated that –although our 138 isolates represented about 80% of the Sal. ruber population– the total population in one saltern pond is composed of 5,500 to 11,000 genomovars, the great majority of which appear to be rare in-situ. These data also revealed that the most frequently recovered isolate in lab media was often not the most abundant genomovar in-situ, suggesting that cultivation biases are significant, even in cases that cultivation procedures are thought to be robust. The methodology and ANI thresholds outlined here should represent a useful guide for future microdiversity surveys of additional microbial species.


