2024-07-24 パシフィック・ノースウェスト国立研究所(PNNL)
<関連情報>
- https://www.pnnl.gov/publications/predicting-metabolic-potential-bacteria-limited-genome-data
- https://elifesciences.org/articles/85749
MetaPathPredictで不完全な細菌ゲノムの代謝モジュールを予測する Predicting metabolic modules in incomplete bacterial genomes with MetaPathPredict
David Geller-McGrath,Kishori M Konwar,Virginia P Edgcomb,Maria Pachiadaki,Jack W Roddy,Travis J Wheeler,Jason E McDermott
eLife Published:May 2, 2024
DOI:https://doi.org/10.7554/eLife.85749
Abstract
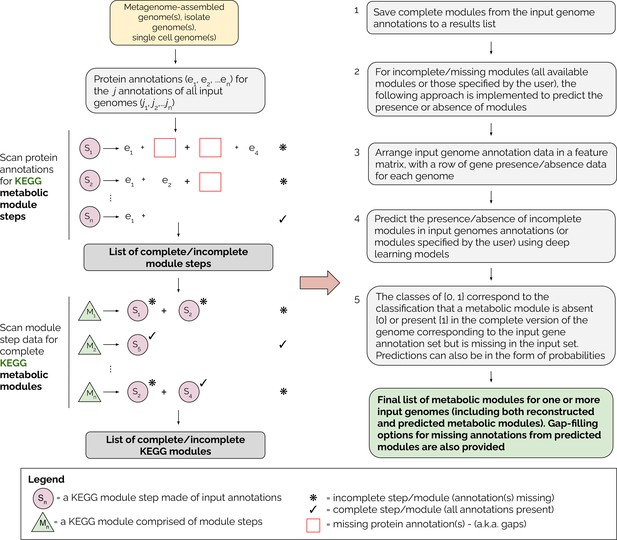
The reconstruction of complete microbial metabolic pathways using ‘omics data from environmental samples remains challenging. Computational pipelines for pathway reconstruction that utilize machine learning methods to predict the presence or absence of KEGG modules in incomplete genomes are lacking. Here, we present MetaPathPredict, a software tool that incorporates machine learning models to predict the presence of complete KEGG modules within bacterial genomic datasets. Using gene annotation data and information from the KEGG module database, MetaPathPredict employs deep learning models to predict the presence of KEGG modules in a genome. MetaPathPredict can be used as a command line tool or as a Python module, and both options are designed to be run locally or on a compute cluster. Benchmarks show that MetaPathPredict makes robust predictions of KEGG module presence within highly incomplete genomes.