2025-03-11 アルゴンヌ国立研究所
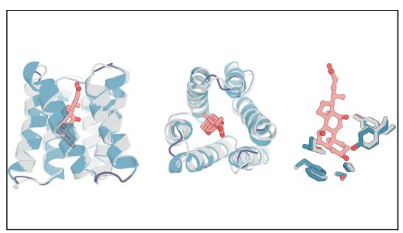
The crystal structure of CHD_r1 (gray) is very similar to the computational design model (colored). (Image by Linna An, et al., Science.)
<関連資料>
- https://www.anl.gov/article/breaking-boundaries-in-biomedicine-advanced-photon-source-enables-protein-design
- https://www.science.org/doi/10.1126/science.adn3780
形状補完擬似環を用いた多様な小分子の結合と検知 Binding and sensing diverse small molecules using shape-complementary pseudocycles
Linna An, Meerit Said, Long Tran, Sagardip Majumder, Inna Goreshnik, Gyu Rie Lee, David Juergens, Justas Dauparas, Ivan Anishchenko, […], and David Baker +15 authorsAuthors Info & Affiliations
Science 18 Jul 2024 Vol 385, Issue 6706 pp. 276-282
DOI: 10.1126/science.adn3780
Editor’s summary
A major current challenge in protein design is the de novo creation of selective and high-affinity small-molecule–binding proteins. One promising concept is a small domain that could be incorporated into other applications with ease. An et al. designed small, pseudocyclic peptides with a central cavity that can be expanded or reduced by changing the number of repeating units to match the size of a target ligand. High-throughput screening and computational resampling enabled the creation of second-generation binders with substantially higher affinities for four example target molecules. The authors demonstrate two applications of such binders that rely on incorporating them as a domain into a larger, de novo–designed protein: a nanopore fusion for small-molecule sensing and a split protein for chemically induced dimerization. —Michael A. Funk
Abstract
We describe an approach for designing high-affinity small molecule–binding proteins poised for downstream sensing. We use deep learning–generated pseudocycles with repeating structural units surrounding central binding pockets with widely varying shapes that depend on the geometry and number of the repeat units. We dock small molecules of interest into the most shape complementary of these pseudocycles, design the interaction surfaces for high binding affinity, and experimentally screen to identify designs with the highest affinity. We obtain binders to four diverse molecules, including the polar and flexible methotrexate and thyroxine. Taking advantage of the modular repeat structure and central binding pockets, we construct chemically induced dimerization systems and low-noise nanopore sensors by splitting designs into domains that reassemble upon ligand addition.