2025-04-09 宮崎大学
<関連情報>
- https://www.miyazaki-u.ac.jp/newsrelease/edu-info/499rna-rnp.html
- https://www.miyazaki-u.ac.jp/public-relations/20250409_01_press.pdf
- https://academic.oup.com/nar/article/53/6/gkaf212/8090312
- https://academic.oup.com/nar/article/50/2/601/6313240
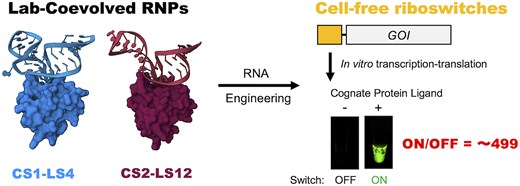
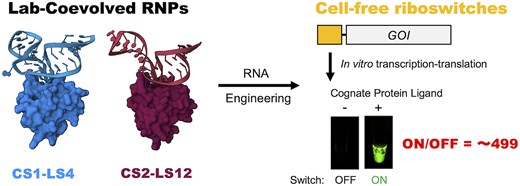
ラボで進化したRNA-RBPペアの構造的洞察と無細胞系における合成リボスイッチの応用 Structural insights into lab-coevolved RNA–RBP pairs and applications of synthetic riboswitches in cell-free system
Keisuke Fukunaga, Takamasa Teramoto, Momoka Nakashima, Toshitaka Ohtani, Riku Katsuki, Tomoaki Matsuura, Yohei Yokobayashi, Yoshimitsu Kakuta
Nucleic Acids Research Published:22 March 2025
DOI:https://doi.org/10.1093/nar/gkaf212
Graphical Abstract
Abstract
CS1–LS4 and CS2–LS12 are ultra-high affinity and orthogonal RNA–protein pairs that were identified by PD-SELEX (Phage Display coupled with Systematic Evolution of Ligands by EXponential enrichment). To investigate the molecular basis of the lab-coevolved RNA–RBP pairs, we determined the structures of the CS1–LS4 and CS2–LS12 complexes and the LS12 homodimer in an RNA-free state by X-ray crystallography. The structural analyses revealed that the lab-coevolved RNA–RBPs have acquired unique molecular recognition mechanisms, whereas the overall structures of the RNP complexes were similar to the typical kink-turn RNA-L7Ae complex. The orthogonal RNA–RBP pairs were applied to construct high-performance cell-free riboswitches that regulate translation in response to LS4 or LS12. In addition, by using the orthogonal protein-responsive switches, we generated an AND logic gate that outputs staphylococcal γ-hemolysin in cell-free system and carried out hemolysis assay and calcein leakage assay using rabbit red blood cells and artificial cells, respectively.
ライブラリ対ライブラリのin vitro選択による直交RNA-RBPペアの指向性進化 Directed evolution of orthogonal RNA–RBP pairs through library-vs-library in vitro selection
Keisuke Fukunaga, Yohei Yokobayashi
Nucleic Acids Research Published:05 July 2021
DOI:https://doi.org/10.1093/nar/gkab527
Graphical Abstract
Library-vs-library selection of RNA and RNA-binding protein (RBP) resulted in orthogonal RNA–RBP pairs with picomolar affinity and high selectivity.
Abstract
RNA-binding proteins (RBPs) and their RNA ligands play many critical roles in gene regulation and RNA processing in cells. They are also useful for various applications in cell biology and synthetic biology. However, re-engineering novel and orthogonal RNA–RBP pairs from natural components remains challenging while such synthetic RNA–RBP pairs could significantly expand the RNA–RBP toolbox for various applications. Here, we report a novel library-vs-library in vitro selection strategy based on Phage Display coupled with Systematic Evolution of Ligands by EXponential enrichment (PD-SELEX). Starting with pools of 1.1 × 1012 unique RNA sequences and 4.0 × 108 unique phage-displayed L7Ae-scaffold (LS) proteins, we selected RNA–RBP complexes through a two-step affinity purification process. After six rounds of library-vs-library selection, the selected RNAs and LS proteins were analyzed by next-generation sequencing (NGS). Further deconvolution of the enriched RNA and LS protein sequences revealed two synthetic and orthogonal RNA–RBP pairs that exhibit picomolar affinity and >4000-fold selectivity.