2025-04-10 イェール大学
<関連情報>
- https://medicine.yale.edu/news-article/regulators-of-dna-folding-could-be-targets-for-treating-cancer/
- https://www.nature.com/articles/s41592-025-02652-z
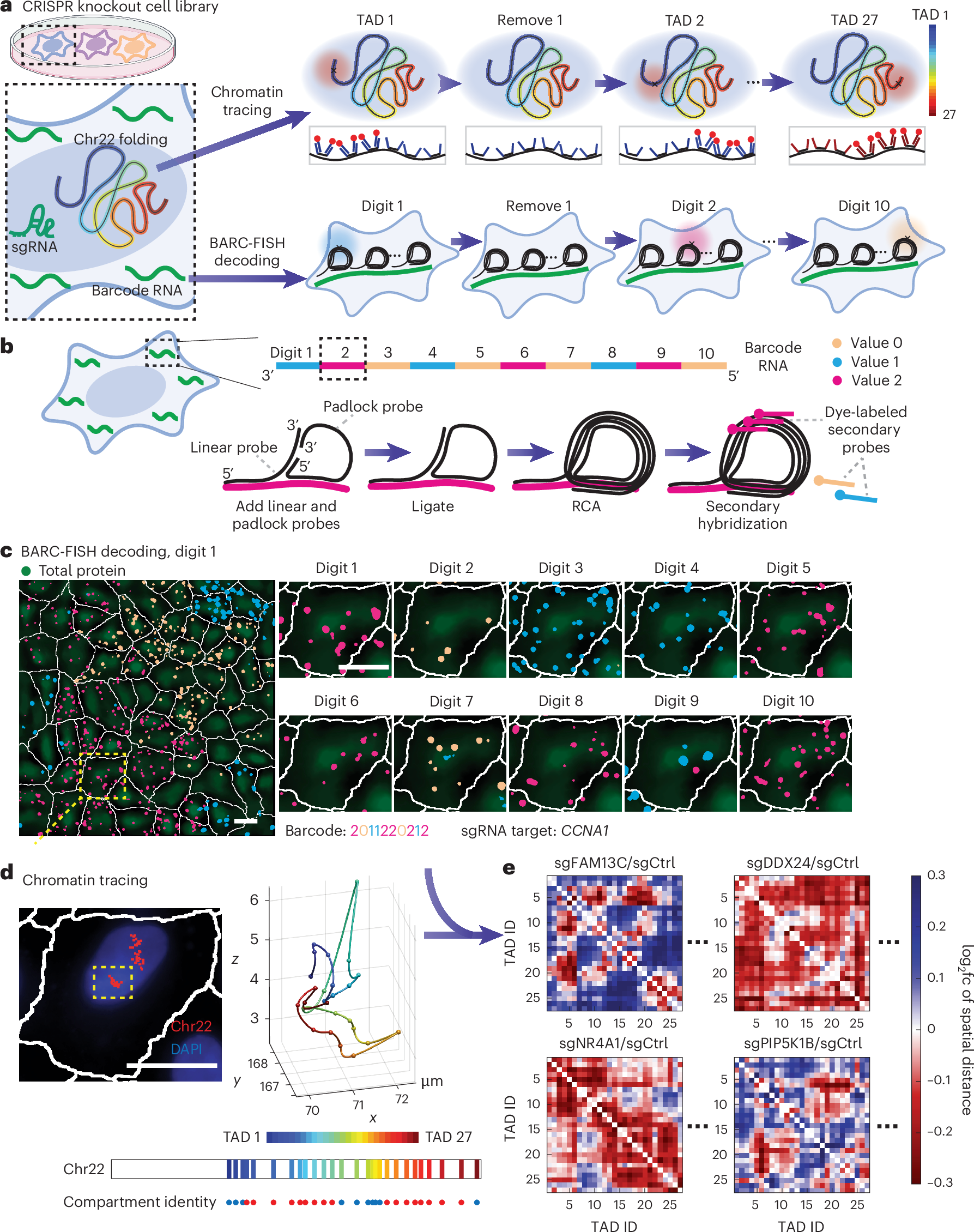
摂動追跡により、マルチスケール3Dゲノム調節因子のハイコンテンツスクリーニングが可能に Perturb-tracing enables high-content screening of multi-scale 3D genome regulators
Yubao Cheng,Mengwei Hu,Bing Yang,Tyler B. Jensen,Yuan Zhang,Tianqi Yang,Ruihuan Yu,Zhaoxia Ma,Jonathan S. D. Radda,Shengyan Jin,Chongzhi Zang & Siyuan Wang
Nature Methods Published:10 April 2025
DOI:https://doi.org/10.1038/s41592-025-02652-z
Abstract
Three-dimensional (3D) genome organization becomes altered during development, aging and disease, but the factors regulating chromatin topology are incompletely understood and currently no technology can efficiently screen for new regulators of multi-scale chromatin organization. Here, we developed an image-based high-content screening platform (Perturb-tracing) that combines pooled CRISPR screens, a cellular barcode readout method (BARC-FISH) and chromatin tracing. We performed a loss-of-function screen in human cells, and visualized alterations to their 3D chromatin folding conformations, alongside perturbation-paired barcode readout in the same single cells. We discovered tens of new regulators of chromatin folding at different length scales, ranging from chromatin domains and compartments to chromosome territory. A subset of the regulators exhibited 3D genome effects associated with loop extrusion and A–B compartmentalization mechanisms, while others were largely unrelated to these known 3D genome mechanisms. Finally, we identified new regulators of nuclear architectures and found a functional link between chromatin compaction and nuclear shape. Altogether, our method enables scalable, high-content identification of chromatin and nuclear topology regulators that will stimulate new insights into the 3D genome.