2025-05-23 大阪大学,Spiber株式会社,科学技術振興機構
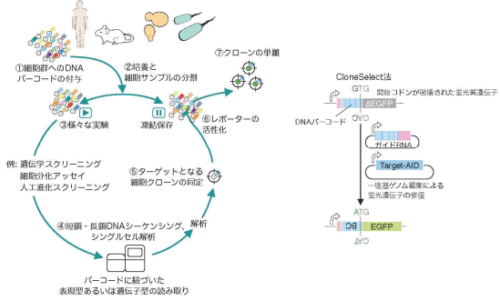
図1:一般的な遡及的クローン単離(左)と CloneSelect 法で用いられた遺伝子回路(右)。
<関連情報>
- https://www.jst.go.jp/pr/announce/20250523/index.html
- https://www.jst.go.jp/pr/announce/20250523/pdf/20250523.pdf
- https://www.nature.com/articles/s41587-025-02649-1
正確なクローン分離のためのマルチドメイン遺伝子バーコードシステム A multi-kingdom genetic barcoding system for precise clone isolation
Soh Ishiguro,Kana Ishida,Rina C. Sakata,Minori Ichiraku,Ren Takimoto,Rina Yogo,Yusuke Kijima,Hideto Mori,Mamoru Tanaka,Samuel King,Shoko Tarumoto,Taro Tsujimura,Omar Bashth,Nanami Masuyama,Arman Adel,Hiromi Toyoshima,Motoaki Seki,Ju Hee Oh,Anne-Sophie Archambault,Keiji Nishida,Akihiko Kondo,Satoru Kuhara,Hiroyuki Aburatani,Ramon I. Klein Geltink,… Nozomu Yachie
Nature Biotechnology Published:21 May 2025
DOI:https://doi.org/10.1038/s41587-025-02649-1
Abstract
Cell-tagging strategies with DNA barcodes have enabled the analysis of clone size dynamics and clone-restricted transcriptomic landscapes in heterogeneous populations. However, isolating a target clone that displays a specific phenotype from a complex population remains challenging. Here we present a multi-kingdom genetic barcoding system, CloneSelect, which enables a target cell clone to be triggered to express a reporter gene for isolation through barcode-specific CRISPR base editing. In CloneSelect, cells are first stably tagged with DNA barcodes and propagated so that their subpopulation can be subjected to a given experiment. A clone that shows a phenotype or genotype of interest at a given time can then be isolated from the initial or subsequent cell pools stored during the experiment using CRISPR base editing. CloneSelect is scalable and compatible with single-cell RNA sequencing. We demonstrate the versatility of CloneSelect in human embryonic kidney 293T cells, mouse embryonic stem cells, human pluripotent stem cells, yeast cells and bacterial cells.