2025-07-04 東京大学
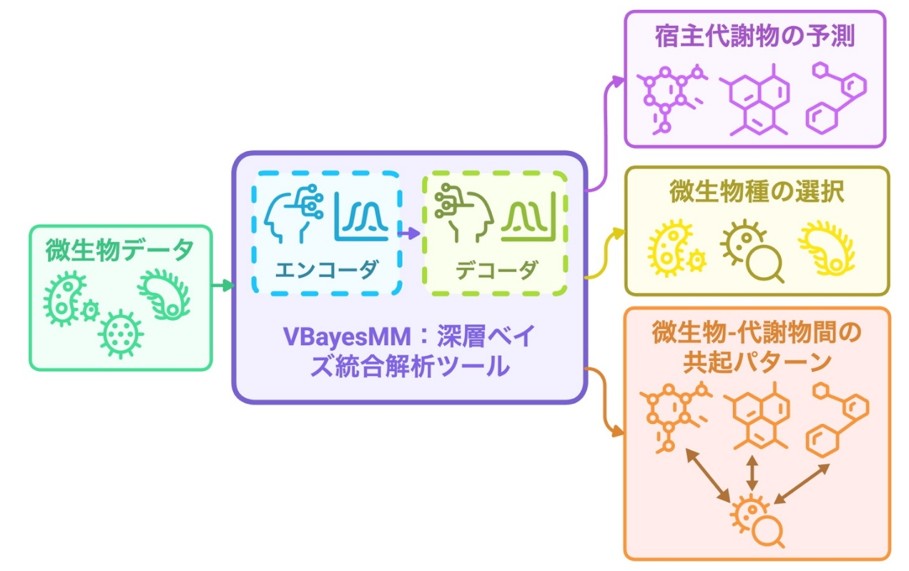
微生物-宿主代謝物間の関連性を発見するためのVBayesMM法
<関連情報>
- https://www.s.u-tokyo.ac.jp/ja/press/10840/
- https://academic.oup.com/bib/article/26/4/bbaf300/8185645?login=false
VBayesMM:高次元マイクロバイオームマルチオミクスデータの重要な関係を優先順位付けする変分ベイズニューラルネットワーク VBayesMM: variational Bayesian neural network to prioritize important relationships of high-dimensional microbiome multiomics data
Tung Dang , Artem Lysenko , Keith A Boroevich , Tatsuhiko Tsunoda
Briefings in Bioinformatics Published:04 July 2025
DOI:https://doi.org/10.1093/bib/bbaf300
Abstract
The analysis of high-dimensional microbiome multiomics datasets is crucial for understanding the complex interactions between microbial communities and host physiological states across health and disease conditions. Despite their importance, current methods, such as the microbe–metabolite vectors approach, often face challenges in predicting metabolite abundances from microbial data and identifying keystone species. This arises from the vast dimensionality of metagenomics data, which complicates the inference of significant relationships, particularly the estimation of co-occurrence probabilities between microbes and metabolites. Here we propose the variational Bayesian microbiome multiomics (VBayesMM) approach, which aims to improve the prediction of metabolite abundances from microbial metagenomics data by incorporating a spike-and-slab prior within a Bayesian neural network. This allows VBayesMM to rapidly and precisely identify crucial microbial species, leading to more accurate estimations of co-occurrence probabilities between microbes and metabolites, while also robustly managing the uncertainty inherent in high-dimensional data. Moreover, we have implemented variational inference to address computational bottlenecks, enabling scalable analysis across extensive multiomics datasets. Our large-scale comparative evaluations demonstrate that VBayesMM not only outperforms existing methods in predicting metabolite abundances but also provides a scalable solution for analyzing massive datasets. VBayesMM enhances the interpretability of the Bayesian neural network by identifying a core set of influential microbial species, thus facilitating a deeper understanding of their probabilistic relationships with the host.

