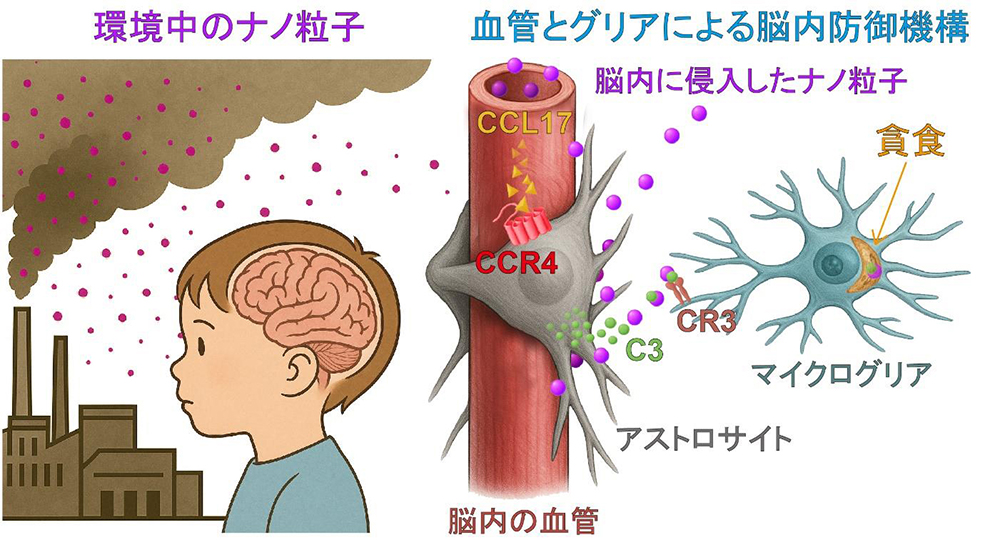
2025-07-17 東京大学
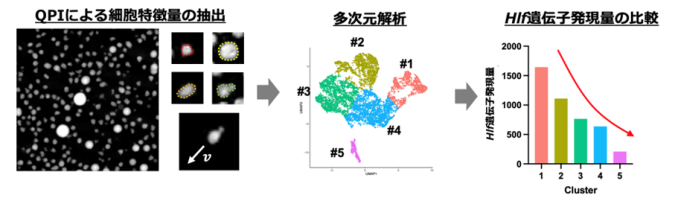
図1:造血幹細胞のHlf遺伝子発現量を、細胞固有の特徴量のみから予測することに成功
HSCに対してQPIにより取得した細胞動態データから特徴量を抽出し、多次元解析を行うことで、細胞群を異なる複数のクラスタに分類可能であることを示した。さらに、同一の特徴量からHlf遺伝子の発現量を予測できることが明らかとなった。
<関連情報>
- https://www.ims.u-tokyo.ac.jp/imsut/jp/about/press/page_00342.html
- https://www.ims.u-tokyo.ac.jp/imsut/content/000011378.pdf
- https://www.nature.com/articles/s41467-025-61846-3
造血幹細胞の多様性を予測する時間動態を用いた定量的位相イメージング Quantitative phase imaging with temporal kinetics predicts hematopoietic stem cell diversity
Takao Yogo,Yuichiro Iwamoto,Hans Jiro Becker,Takaharu Kimura,Reiko Ishida,Ayano Sugiyama-Finnis,Tomomasa Yokomizo,Toshio Suda,Sadao Ota & Satoshi Yamazaki
Nature Communications Published:14 July 2025
DOI:https://doi.org/10.1038/s41467-025-61846-3
Abstract
Innovative identification technologies for hematopoietic stem cells (HSCs) have expanded the scope of stem cell biology. Clinically, the functional quality of HSCs critically influences the safety and therapeutic efficacy of stem cell therapies. However, most analytical techniques capture only a single snapshot, disregarding the temporal context. A comprehensive understanding of the temporal heterogeneity of HSCs necessitates live-cell, real-time and non-invasive analysis. Here, we developed a prediction system for HSC diversity by integrating single-HSC ex vivo expansion technology with quantitative phase imaging (QPI)-driven machine learning. By analyzing the cellular kinetics of individual HSCs, we discovered previously undetectable diversity that snapshot analysis cannot resolve. The QPI-driven algorithm quantitatively evaluates stemness at the single-cell level and leverages temporal information to significantly improve prediction accuracy. This platform advances the field from snapshot-based identification of HSCs to dynamic, time-resolved prediction of their functional quality based on past cellular kinetics.