2025-08-20 ハーバード大学
Web要約 の発言:
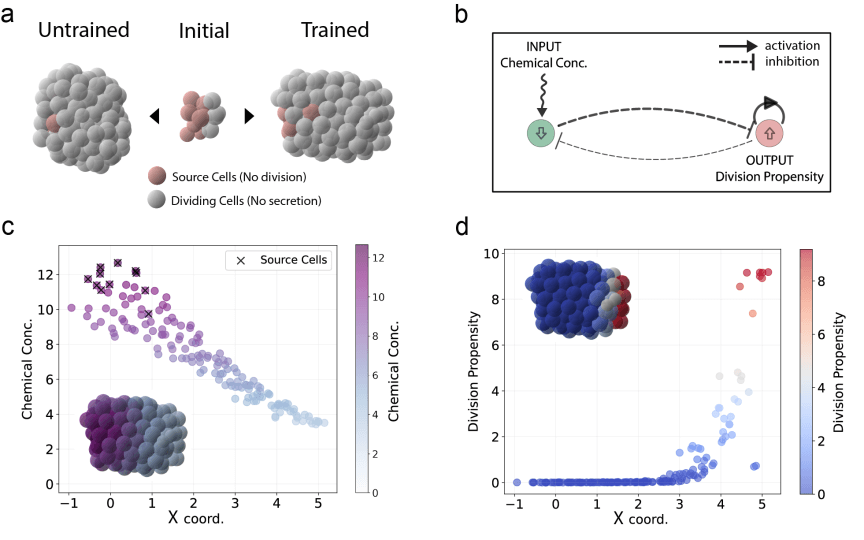 Schematic of horizontal elongation in an optimized cell cluster. (a) Left: the final configuration of a simulation with randomly initialized parameters; right: the final simulation state after learning. Source cells, in red, secrete the growth factor and cannot divide. Proliferating cells, in gray, sense the growth factor and divide in response to it. (b) The learned gene network. The receptor gene is activated only by the presence of the external chemical factor, which results in repression of the division propensity. (c) Chemical gradient created by source cells along the cluster x-coordinate. (d) Division propensity distribution at the end of the simulation along the x-axis, highlighting the concentration of dividing cells at the tip.
Schematic of horizontal elongation in an optimized cell cluster. (a) Left: the final configuration of a simulation with randomly initialized parameters; right: the final simulation state after learning. Source cells, in red, secrete the growth factor and cannot divide. Proliferating cells, in gray, sense the growth factor and divide in response to it. (b) The learned gene network. The receptor gene is activated only by the presence of the external chemical factor, which results in repression of the division propensity. (c) Chemical gradient created by source cells along the cluster x-coordinate. (d) Division propensity distribution at the end of the simulation along the x-axis, highlighting the concentration of dividing cells at the tip.
<関連情報>
- https://seas.harvard.edu/news/2025/08/optimizing-how-cells-self-organize
- https://www.nature.com/articles/s43588-025-00851-4
細胞クラスターの形態形成を微分可能プログラミングで制御する Engineering morphogenesis of cell clusters with differentiable programming
Ramya Deshpande,Francesco Mottes,Ariana-Dalia Vlad,Michael P. Brenner & Alma Dal Co
Nature Computational Science Published:13 August 2025
DOI:https://doi.org/10.1038/s43588-025-00851-4
Abstract
Understanding the fundamental rules of organismal development is a central, unsolved problem in biology. These rules dictate how individual cellular actions coordinate over macroscopic numbers of cells to grow complex structures with exquisite functionality. We use recent advances in automatic differentiation to discover local interaction rules and genetic networks that yield emergent, systems-level characteristics in a model of development. We consider a growing tissue with cellular interactions mediated by morphogen diffusion, cell adhesion and mechanical stress. Each cell has an internal genetic network that is used to make decisions based on the cell’s local environment. Here we show that one can learn the parameters governing cell interactions in the form of interpretable genetic networks for complex developmental scenarios. When combined with recent experimental advances measuring spatio-temporal dynamics and gene expression of cells in a growing tissue, the methodology outlined here offers a promising path to unraveling the cellular bases of development.