2025-09-08 ペンシルベニア州立大学(PennState)
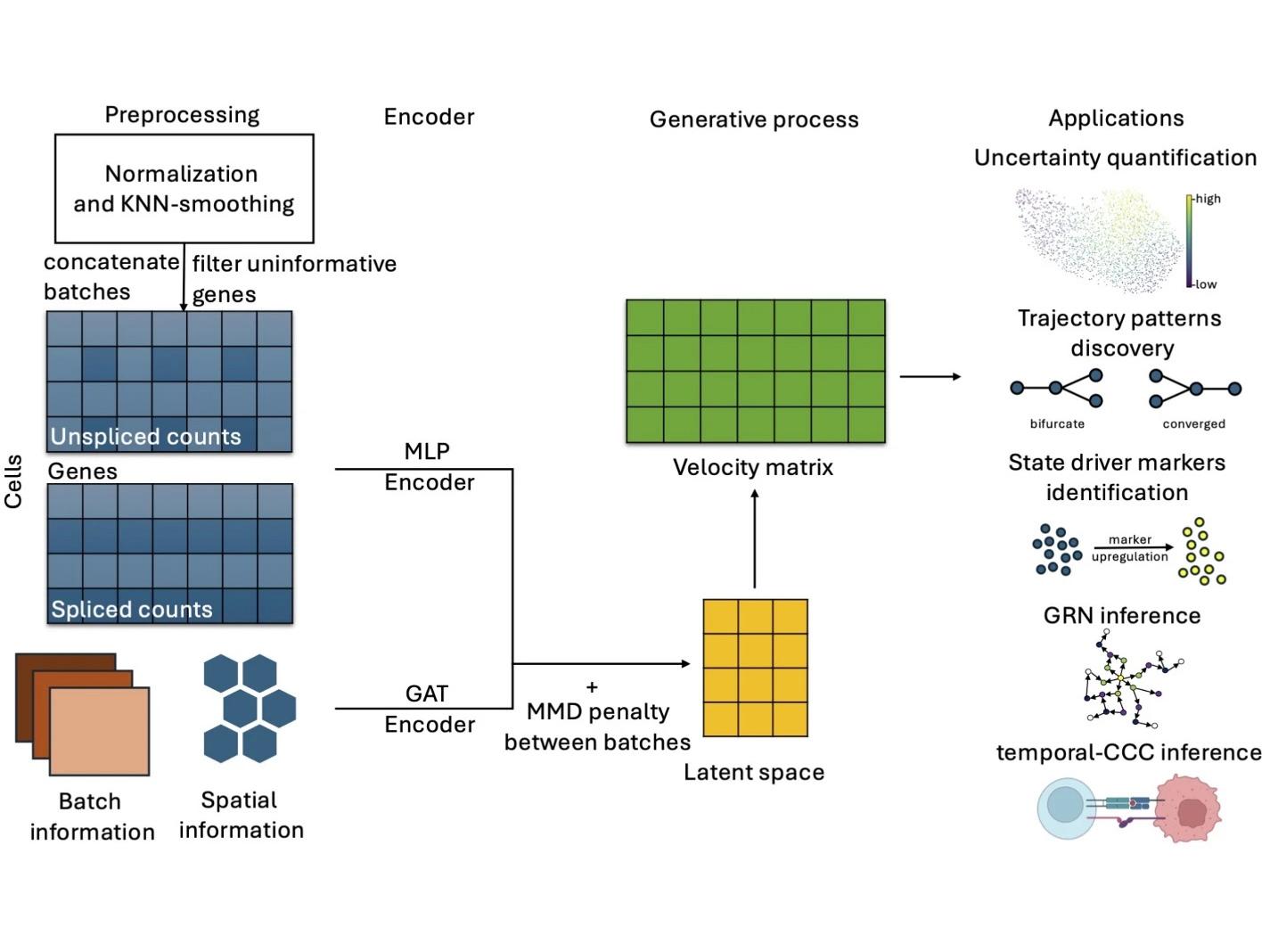
A new method to measure RNA velocity — which describes rate of gene expression in a single cell — overcomes limitations of previous methods and can incorporate spatial information as well as data from cells processed at different times. The new method, called spVelo incorporates two types of neural networks: a Variational Autoencoder that models gene expression and a Graph Attention Network that allows the incorporation of spatial and batch information from the sequencing data. Credit: Provided by the researchers. All Rights Reserved.
<関連情報>
- https://www.psu.edu/news/eberly-college-science/story/new-method-calculates-rate-gene-expression-understand-cell-fate
- https://genomebiology.biomedcentral.com/articles/10.1186/s13059-025-03701-8
spVelo:マルチバッチ空間トランスクリプトームデータ向けRNA速度推定法 spVelo: RNA velocity inference for multi-batch spatial transcriptomics data
Wenxin Long,Tianyu Liu,Lingzhou Xue & Hongyu Zhao
Genome Biology Published:11 August 2025
DOI:https://doi.org/10.1186/s13059-025-03701-8
Abstract
RNA velocity has emerged as a powerful tool to interpret transcriptional dynamics and infer trajectory from snapshot datasets. However, current methods fail to utilize the spatial information inherent in spatial transcriptomics and lack scalability in multi-batch datasets. Here, we introduce spVelo, a scalable framework for RNA velocity inference of multi-batch spatial transcriptomics data. spVelo supports several downstream applications, including uncertainty quantification, complex trajectory pattern discovery, driver marker identification, gene regulatory network inference, and temporal cell-cell communication inference. spVelo has the potential to provide deeper insights into complex tissue organization and underscore biological mechanisms based on spatially resolved patterns.