2025-09-18 カリフォルニア大学サンディエゴ校(UCSD)
<関連情報>
- https://today.ucsd.edu/story/small-nuclear-rna-base-editing-a-safer-alternative-to-crispr-uc-san-diego-researchers-find
- https://www.nature.com/articles/s41589-025-02026-8
小核RNAを用いた哺乳類転写産物におけるRNA塩基編集の効率化 Enhancing RNA base editing on mammalian transcripts with small nuclear RNAs
Aaron A. Smargon,Deepak Pant,Trent A. Gomberg,Christian Fagre,Sofia Glynne,Johnathan Nguyen,Jack T. Naritomi,Wendy V. Gilbert & Gene W. Yeo
Nature Chemical Biology Published:18 September 2025
DOI:https://doi.org/10.1038/s41589-025-02026-8
Abstract
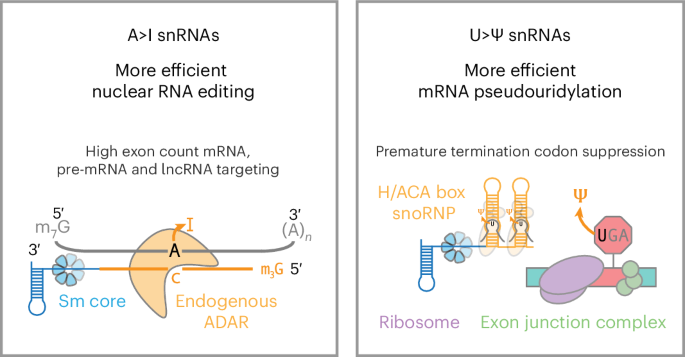
Endogenous uridine-rich small nuclear RNAs (U snRNAs) form RNA–protein complexes to process eukaryotic pre-mRNA into mRNA. Previous studies have demonstrated programmable U snRNA guide-targeted exon inclusion and exclusion. Here we investigated whether snRNAs can also enhance RNA base editing over state-of-the-art RNA-targeting technologies in human cells. Compared with adenosine deaminase acting on RNA (ADAR)-recruiting circular RNAs, we find that guided A>I snRNAs consistently increase adenosine-to-inosine editing for higher exon count genes, perturb substantially fewer off-target genes and localize more persistently to the nucleus where ADAR is expressed. A>I snRNAs also more efficiently edit long noncoding RNAs and pre-mRNA 3′ splice sites to promote splicing changes. Lastly, snRNA–H/ACA box snoRNA fusions (U>Ψ snRNAs) increase targeted RNA pseudouridylation without DKC1 overexpression, facilitating improved CFTR rescue from nonsense-mediated mRNA decay in a cystic fibrosis human bronchial epithelial cell model. Our results advance the endogenous protein-mediated RNA base editing toolbox and RNA-targeting technologies to treat genetic diseases.