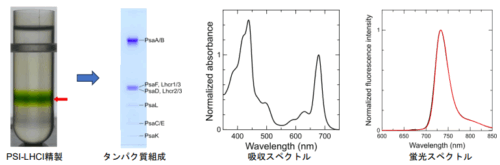
2025-09-23 理化学研究所,東京大学,株式会社ユーグレナ,山形大学,鶴岡工業高等専門学校,高知大学,長崎大学,横浜市立大学

真核生物の遺伝子発現とユーグレナゲノムの非従来型イントロン
(Created in BioRender. Mochida, K. (2025) )
<関連情報>
- https://www.riken.jp/press/2025/20250923_1/index.html
- https://www.pnas.org/doi/10.1073/pnas.2509937122
非従来型イントロンの遺伝学的解析により、ユーグレナにおける共優性非標準スプライシングコードが明らかになる Genetic dissection of nonconventional introns reveals codominant noncanonical splicing code in Euglena
Toshihisa Nomura, June-Sik Kim, Osamu Iwata, +11 , and Keiichi Mochida
Proceedings of the National Academy of Sciences Published:September 23, 2025
DOI:https://doi.org/10.1073/pnas.2509937122
Significance
While eukaryotic genes typically contain introns with conventional GT-AG boundaries, many nonconventional introns, which lack these boundaries, remain poorly characterized. Our genome-wide analysis revealed that, uniquely among eukaryotes, Euglena codominantly employs nonconventional introns. By systematically testing both natural and synthetic introns, we identified key DNA sequence signatures for this unusual splicing mechanism and validated the underlying splicing code. The identification of a prevalent nonconventional splicing code in Euglena expands our understanding of the diversity of RNA processing and opens avenues for exploring noncanonical splicing machineries, with implications for evolutionary biology and biotechnology.
Abstract
Pre-mRNA splicing is essential for eukaryotic gene expression and is achieved through the accurate recognition of exon–intron boundaries. Although nonconventional introns, which do not follow the conventional GT-AG splicing rule, have been identified in several species, these introns are typically rare in any given genome. Here, we demonstrate the widespread occurrence of nonconventional introns (71.8% of all introns) in the Euglena agilis genome and identify consensus motifs at these nonconventional exon–intron boundaries. We assessed the splicing efficiency of nonconventional introns and variants with point mutations via genomic knock-in within the second exon of Glucan synthase-like 2 in Euglena gracilis and genetically defined the sequence signature (5′-N3CDG-/-CH′GN5–6|Rexon-3′) required for their proper splicing. This signature is present in 61.2% of all nonconventional introns detected in the E. agilis genome. Accordingly, we present a noncanonical splicing code for Euglena introns, highlighting the global coexistence of dual splicing rules for conventional and nonconventional introns.