2025-10-20 カリフォルニア工科大学(Caltech)
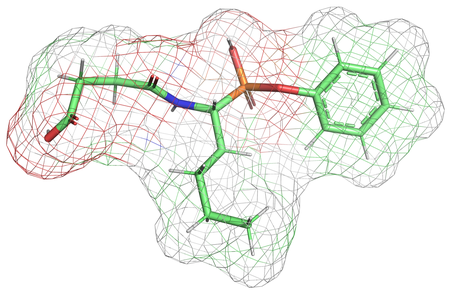
This illustration shows the mesh of anchoring points the team obtained by discretizing the manifold, an estimation of the distribution of atoms and the probable locations of electrons in the molecule. This is important because, as the authors note in the new paper, treating atoms as solid points “does not fully reflect the spatial extent that real atoms occupy in three-dimensional space.”Credit: Liu et al./PNAS
<関連情報>
- https://www.caltech.edu/about/news/new-ai-model-for-drug-design-brings-more-physics-to-bear-in-predictions
- https://www.pnas.org/doi/10.1073/pnas.2415666122
構造に基づく医薬品設計のための多様体制約核レベルノイズ除去拡散モデル Manifold-constrained nucleus-level denoising diffusion model for structure-based drug design
Shengchao Liu, Liang Yan, Weitao Du, +5 , and Anima Anandkumar
Proceedings of the National Academy of Sciences Published:October 6, 2025
DOI:https://doi.org/10.1073/pnas.2415666122
Significance
Structure-based drug design is a critical field in chemistry and biology, and recent advancements in machine learning have significantly enhanced the generation of ligands with high binding affinities for target proteins. However, existing machine learning methods have been overlooking a crucial physical prior: Atoms must maintain a minimum pairwise distance to avoid atomic collisions. We address this issue by introducing a learning approach that incorporates motivated geometric constraints to improve the physical plausibility and binding affinity of generated molecular structures. Our results demonstrate improved molecular realism and practical benefits for drug design tasks.
Abstract
AI models have shown great potential in structure-based drug design, generating ligands with high binding affinities. However, existing models have often overlooked a crucial physical prior: Atoms must maintain a minimum pairwise distance to avoid atomic collision, a phenomenon governed by the balance of attractive and repulsive forces. To mitigate such atomic collisions, we propose NucleusDiff. It enforces spatial distance constraints between atomic nuclei and auxiliary mesh points placed on a spherical surface around each atom, approximating van der Waals boundaries to reduce atomic collisions. We quantitatively evaluate NucleusDiff using the CrossDocked2020 dataset and a COVID-19 therapeutic target, demonstrating that NucleusDiff reduces collision rate by up to 100.00% and enhances binding affinity by up to 22.16%, surpassing state-of-the-art models for structure-based drug design. We also provide qualitative analysis through manifold sampling, visually confirming the effectiveness of NucleusDiff in reducing atomic collisions and improving binding affinities.