2025-11-21 マウントサイナイ医療システム(MSHS)
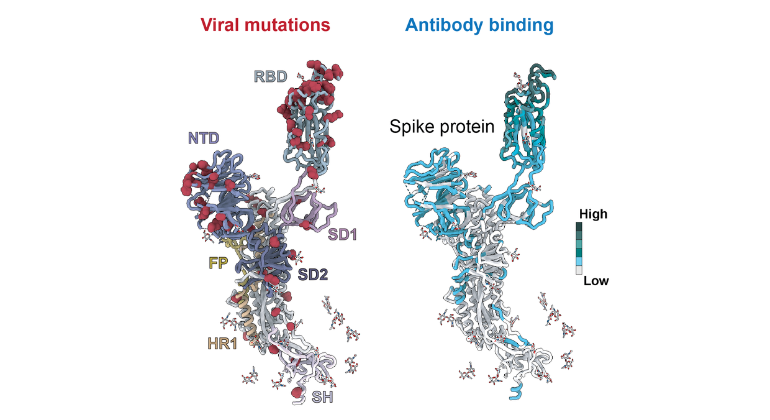
Above: Cartoon model of the SARS-CoV-2 spike protein showing the different regions (or domains) that are associated with known viral mutations and antibodies can recognize and attach to. These include areas involved in binding to human cells and in helping the virus fuse with them. Image credit: Feng, et al., Cell Systems.
<関連情報>
- https://www.mountsinai.org/about/newsroom/2025/scientists-uncover-how-covid-19-variants-outsmart-the-immune-system
- https://www.cell.com/cell-systems/abstract/S2405-4712(25)00285-6
1000のSARS-CoV-2抗体構造が収束結合とほぼ普遍的な免疫回避を明らかに One thousand SARS-CoV-2 antibody structures reveal convergent binding and near-universal immune escape
Zirui Feng ∙ Zhe Sang ∙ Yufei Xiang ∙ … ∙ Dina Schneidman-Duhovny ∙ Adolfo García-Sastre ∙ Yi Shi
Cell Systems Published:November 21, 2025
DOI:https://doi.org/10.1016/j.cels.2025.101452
Highlights
- Structural atlas of >1,100 SARS-CoV-2 antibody complexes (16% of all antibody structures)
- 99% of receptor-binding domain surface residues targeted by antibodies
- >96% of epitopes and 1/3 of epitope residues mutated; only rare survivors retain binding
- Surviving nanobodies bind highly conserved sites yet show limited neutralization
Summary
Understanding antibody recognition and adaptation to viral evolution is central to vaccine and therapeutic development. Over 1,100 SARS-CoV-2 antibody structures have been resolved, marking the largest structural biology effort for a single pathogen. We present a comprehensive analysis of this landmark dataset to investigate the principles of antibody recognition and immune escape. Human immunoglobulins and camelid single-chain antibodies dominate, collectively mapping 99% of the receptor-binding domain. Despite remarkable sequence and conformational diversity, antibodies exhibit convergence in their paratope structures, revealing evolutionary constraints in epitope selection. Analyses reveal near-universal immune escape of antibodies, including all clinical monoclonals, by advanced variants such as KP3.1.1. On average, over one-third of antibody epitope residues are mutated. These findings support pervasive immune escape, underscoring the need to effectively leverage multi-epitope-targeting strategies to achieve durable immunity. To support community accessibility, we developed an interactive web server for visualization and analysis of antibody-antigen complexes and mutational data.