2025-11-24 カリフォルニア大学リバーサイド校(UCR)
<関連情報>
- https://news.ucr.edu/articles/2025/11/24/new-clues-why-some-animals-live-longer
- https://www.nature.com/articles/s41467-025-65339-1
選択的スプライシング制御が寿命の最大化に及ぼす影響 The implications of alternative splicing regulation for maximum lifespan
Wei Jiang,Sika Zheng & Liang Chen
Nature Communications Published:24 November 2025
DOI:https://doi.org/10.1038/s41467-025-65339-1
Abstract
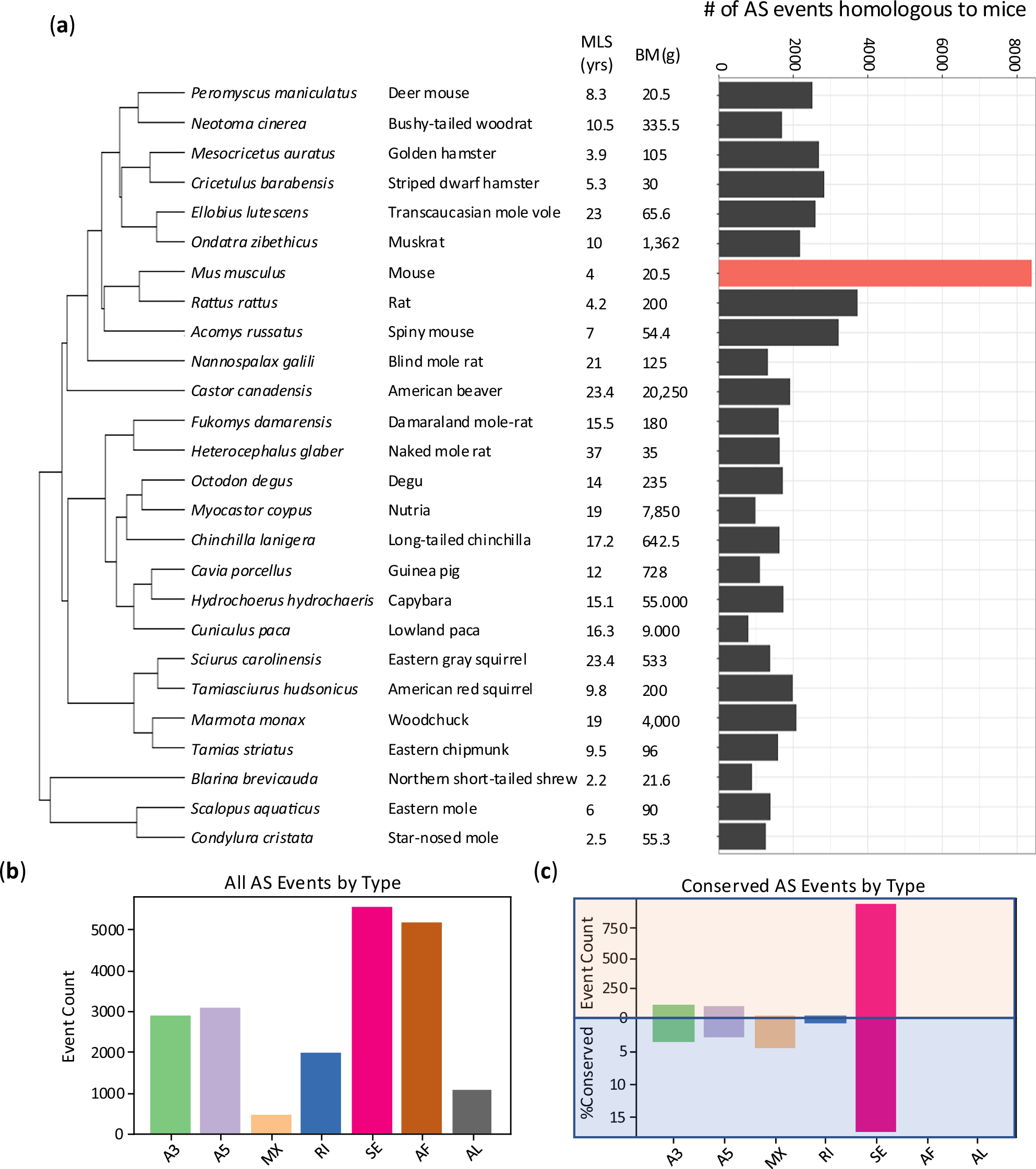
Mammalian maximum lifespan (MLS) varies over a hundred-fold, yet the molecular mechanisms underlying this diversity remain unclear. We present a cross-species analysis of alternative splicing (AS) across six tissues in 26 mammals, identifying hundreds of conserved AS events significantly associated with MLS, with the brain containing twice as many tissue-specific events as peripheral tissues. MLS-AS events are enriched in pathways related to mRNA processing, stress response, neuronal functions, and epigenetic regulation, and are largely distinct from genes whose expression correlates with MLS, indicating that AS captures unique lifespan-related signals. The brain exhibits certain associations divergent from peripheral tissues and reduced overlap with body mass (BM)-associated splicing; neither is observed at the gene expression level. While MLS- and age-associated AS events show limited overlap, the shared events are enriched in intrinsically disordered protein regions, suggesting a role in protein flexibility and stress adaptability. Furthermore, MLS-associated AS events display stronger RNA-binding protein (RBP) motif coordination than age-associated ones, highlighting a more genetically programmed adaptation for lifespan determination, in contrast to the more variable splicing changes seen with chronological aging. These findings suggest alternative splicing as a distinct, transcription-independent axis of lifespan regulation, offering insights into the molecular basis of longevity.