2026-02-12 東北大学
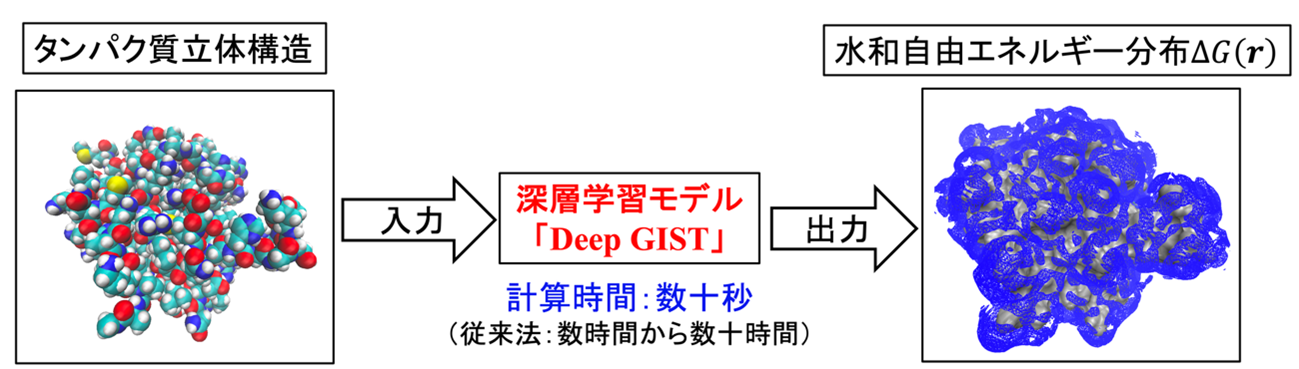
図1. Deep GISTの概要。タンパク質の立体構造を入力すると、その周囲の水和自由エネルギー分布(青色部分)を深層学習モデルにより高速に予測・出力する。
<関連情報>
- https://www.tohoku.ac.jp/japanese/2026/02/press20260212-02-deep.html
- https://pubs.acs.org/doi/10.1021/acs.jcim.5c02389
Deep GIST: タンパク質周囲の水和熱力学分布を予測するためのディープラーニングモデル Deep GIST: Deep Learning Models for Predicting the Distribution of Hydration Thermodynamics around Proteins
Yusaku Fukushima,and Takashi Yoshidome
Journal of Chemical Information and Modeling Published: January 23, 2026
DOI:https://doi.org/10.1021/acs.jcim.5c02389
Abstract
Hydration thermodynamic quantities are essential for understanding protein function from a free-energy perspective. The grid inhomogeneous solvation theory (GIST) enables the computation of spatial distributions of hydration energy, ΔEW(r), and hydration entropy, ΔSW(r), using molecular dynamics (MD) simulations, from which the distribution of the hydration free energy, ΔGW(r), is obtained as ΔGW(r) = ΔEW(r) – TΔSW(r), where T is the absolute temperature. However, GIST is computationally demanding, requiring tens of hours to compute these distributions. To overcome this bottleneck, we developed a set of deep learning models capable of predicting ΔEW(r), TΔSW(r), and ΔGW(r). Our deep learning models completed these predictions within tens of seconds using a single graphics processing unit. The resulting distributions achieved coefficient of determination values of 0.76–0.84 for ΔGW(r) when compared to GIST results, and lower values were obtained for ΔEW(r) and TΔSW(r). As a practical application, we examined the free energy change required for a water molecule to move from the bulk region to the ligand-binding site, ΔGW,replace, using both our deep learning model and GIST. A high correlation coefficient of 0.78 was observed between the predictions of our model and GIST, confirming its reliability. Furthermore, the results for a representative protein were consistent with experimental data of the corresponding protein–ligand complex: Water molecules with low ΔGW,replace values located near crystallographic waters, suggesting retention upon ligand binding, whereas those with unfavorable values overlapped with the ligand, indicating displacement upon the ligand binding. These findings demonstrate that our deep learning models provide an efficient and accurate alternative to GIST for predicting hydration thermodynamics and enable the consideration of protein conformational fluctuations, which is difficult to achieve with conventional GIST. The program called “Deep GIST” is available under the GNU General Public License from https://github.com/YoshidomeGroup-Hydration/Deep-GIST.


