有害な細菌やウイルスを阻止するナノマシン薬が登場する可能性 Nanoengineered drugs that stop harmful bacteria and viruses could be on the horizon
2022-05-16 ミシガン大学
ミシガン大学で開発された、ナノ粒子とタンパク質の相互作用を予測する新しい機械学習モデルは、その実現に一歩近づいたと言えるでしょう。
新しい機械学習アルゴリズムが、ナノ粒子を3つの異なる方法で記述して、タンパク質と比較しています。1つ目は従来の化学的な記述でしたが、構造に関わる2つは、ナノ粒子が特定のタンパク質とロックアンドキーで一致するかどうかを予測するために最も重要であることが判明しました。
<関連情報>
- https://news.umich.edu/nanobiotics-model-predicts-how-nanoparticles-interact-with-proteins/
- https://www.nature.com/articles/s43588-022-00229-w
生体および生体模倣ナノスケール複合体の統一的な構造記述子Unifying structural descriptors for biological and bioinspired nanoscale complexes
Minjeong Cha,Emine Sumeyra Turali Emre,Xiongye Xiao,Ji-Young Kim,Paul Bogdan,J. Scott VanEpps,Angela Violi & Nicholas A. Kotov
Nature Computational Science Published:28 April 2022
DOI:https://doi.org/10.1038/s43588-022-00229-w
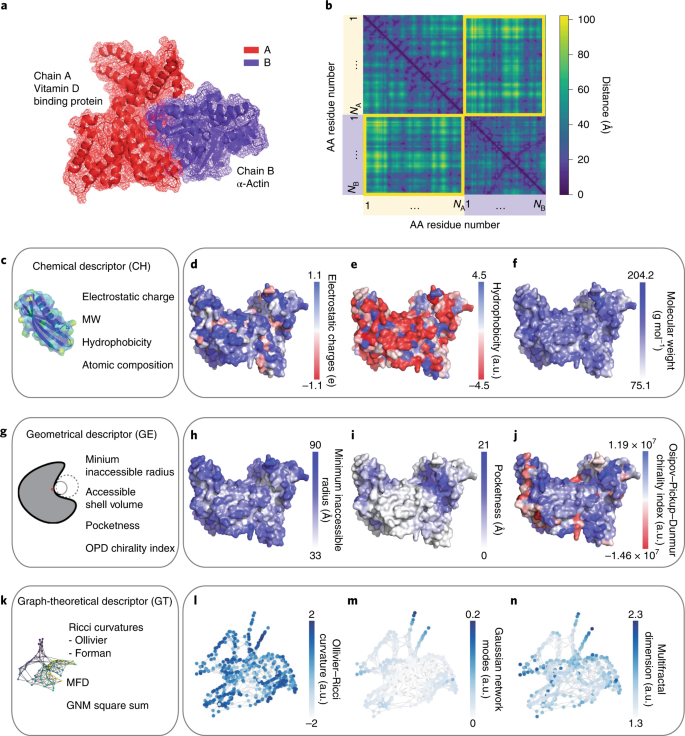
Abstract
Biomimetic nanoparticles are known to serve as nanoscale adjuvants, enzyme mimics and amyloid fibrillation inhibitors. Their further development requires better understanding of their interactions with proteins. The abundant knowledge about protein–protein interactions can serve as a guide for designing protein–nanoparticle assemblies, but the chemical and biological inputs used in computational packages for protein–protein interactions are not applicable to inorganic nanoparticles. Analysing chemical, geometrical and graph-theoretical descriptors for protein complexes, we found that geometrical and graph-theoretical descriptors are uniformly applicable to biological and inorganic nanostructures and can predict interaction sites in protein pairs with accuracy >80% and classification probability ~90%. We extended the machine-learning algorithms trained on protein–protein interactions to inorganic nanoparticles and found a nearly exact match between experimental and predicted interaction sites with proteins. These findings can be extended to other organic and inorganic nanoparticles to predict their assemblies with biomolecules and other chemical structures forming lock-and-key complexes.


