2022-07-19 スイス連邦工科大学ローザンヌ校(EPFL)
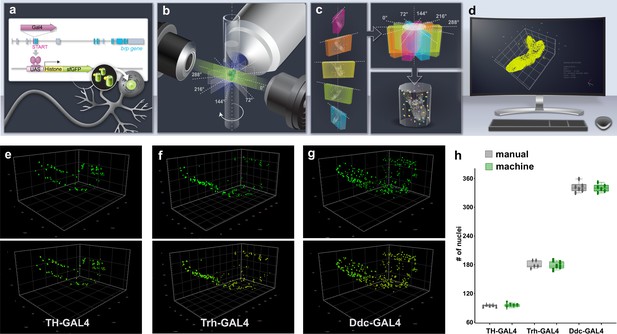
今回、EPFLの研究者らは、神経科学の重要なモデル生物であるショウジョウバエの幼虫の無傷の脳において、超人的な精度と速度で細胞を特定する新しい手法を開発しました。
その結果、これまで考えられていたよりも神経細胞が少なく、グリア(神経細胞を支え保護する細胞)が多いことが判明しました。また、幼虫のオスとメスの脳には予想外の違いがあり、メスの方がオスに比べて神経細胞が大幅に多いことも判明しました。
データの形状を研究するトポロジーと呼ばれる数学の分野の手法を用いてデータを解析したところ、脳のトポロジーだけでその動物の性別を99%の精度で予測することができた.
<関連情報>
- https://actu.epfl.ch/news/computer-genetic-metrics-of-fly-brain-reveal-sex-d/
- https://elifesciences.org/articles/74968
ショウジョウバエの中枢神経系の細胞定量から明らかになった性的二型性 Intact Drosophila central nervous system cellular quantitation reveals sexual dimorphism
Wei Jiao,Gard Spreemann,Evelyne Ruchti,Soumya Banerjee,Samuel Vernon,Ying Shi,R Steven Stowers,Kathryn Hess,Brian D McCabe
eLife Published:Jul 8, 2022
DOI:https://doi.org/10.7554/eLife.74968
Abstract
Establishing with precision the quantity and identity of the cell types of the brain is a prerequisite for a detailed compendium of gene and protein expression in the central nervous system (CNS). Currently, however, strict quantitation of cell numbers has been achieved only for the nervous system of Caenorhabditis elegans. Here, we describe the development of a synergistic pipeline of molecular genetic, imaging, and computational technologies designed to allow high-throughput, precise quantitation with cellular resolution of reporters of gene expression in intact whole tissues with complex cellular constitutions such as the brain. We have deployed the approach to determine with exactitude the number of functional neurons and glia in the entire intact larval Drosophila CNS, revealing fewer neurons and more glial cells than previously predicted. We also discover an unexpected divergence between the sexes at this juvenile developmental stage, with the female CNS having significantly more neurons than that of males. Topological analysis of our data establishes that this sexual dimorphism extends to deeper features of CNS organisation. We additionally extended our analysis to quantitate the expression of voltage-gated potassium channel family genes throughout the CNS and uncover substantial differences in abundance. Our methodology enables robust and accurate quantification of the number and positioning of cells within intact organs, facilitating sophisticated analysis of cellular identity, diversity, and gene expression characteristics.