2024-08-22 ノースカロライナ州立大学(NCState)
<関連情報>
- https://news.ncsu.edu/2024/08/functional-dna-computing/
- https://www.nature.com/articles/s41565-024-01771-6
原始的なDNA記憶装置と計算処理エンジン A primordial DNA store and compute engine
Kevin N. Lin,Kevin Volkel,Cyrus Cao,Paul W. Hook,Rachel E. Polak,Andrew S. Clark,Adriana San Miguel,Winston Timp,James M. Tuck,Orlin D. Velev & Albert J. Keung
Nature Nanotechnology Published:22 August 2024
DOI:https://doi.org/10.1038/s41565-024-01771-6
Abstract
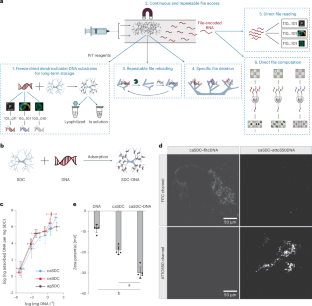
Any modern information system is expected to feature a set of primordial features and functions: a substrate stably carrying data; the ability to repeatedly write, read, erase, reload and compute on specific data from that substrate; and the overall ability to execute such functions in a seamless and programmable manner. For nascent molecular information technologies, proof-of-principle realization of this set of primordial capabilities would advance the vision for their continued development. Here we present a DNA-based store and compute engine that captures these primordial capabilities. This system comprises multiple image files encoded into DNA and adsorbed onto ~50-μm-diameter, highly porous, hierarchically branched, colloidal substrate particles comprised of naturally abundant cellulose acetate. Their surface areas are over 200 cm2 mg-1 with binding capacities of over 1012 DNA oligos mg-1, 10 TB mg-1 or 104 TB cm–3. This ‘dendricolloid’ stably holds DNA files better than bare DNA with an extrapolated ability to be repeatedly lyophilized and rehydrated over 170 times compared with 60 times, respectively. Accelerated ageing studies project half-lives of ~6,000 and 2 million years at 4 °C and -18 °C, respectively. The data can also be erased and replaced, and non-destructive file access is achieved through transcribing from distinct synthetic promoters. The resultant RNA molecules can be directly read via nanopore sequencing and can also be enzymatically computed to solve simplified 3 × 3 chess and sudoku problems. Our study establishes a feasible route for utilizing the high information density and parallel computational advantages of nucleic acids.