2024-09-25 テキサス大学オースチン校(UT Austin)
<関連情報>
- https://news.utexas.edu/2024/09/25/ai-trained-on-evolutions-playbook-develops-proteins-that-spur-drug-and-scientific-discovery/
- https://www.nature.com/articles/s41467-024-49780-2
安定性オラクル:安定化変異を同定するための構造ベースのグラフ変換フレームワーク Stability Oracle: a structure-based graph-transformer framework for identifying stabilizing mutations
Daniel J. Diaz,Chengyue Gong,Jeffrey Ouyang-Zhang,James M. Loy,Jordan Wells,David Yang,Andrew D. Ellington,Alexandros G. Dimakis & Adam R. Klivans
Nature Communications Published:23 July 2024
DOI:https://doi.org/10.1038/s41467-024-49780-2
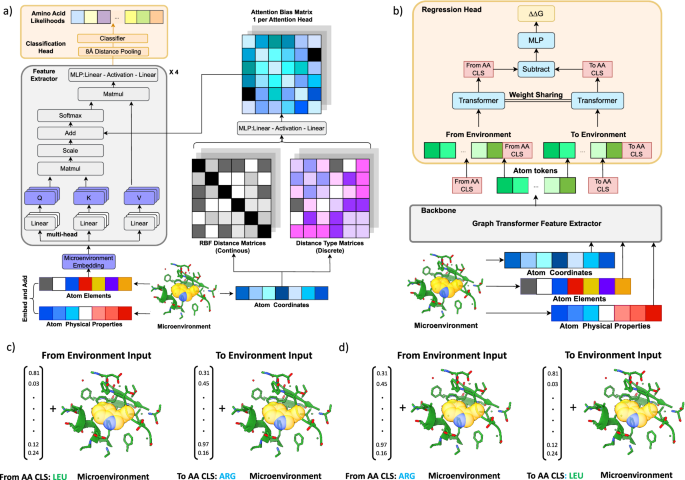
Abstract
Engineering stabilized proteins is a fundamental challenge in the development of industrial and pharmaceutical biotechnologies. We present Stability Oracle: a structure-based graph-transformer framework that achieves SOTA performance on accurately identifying thermodynamically stabilizing mutations. Our framework introduces several innovations to overcome well-known challenges in data scarcity and bias, generalization, and computation time, such as: Thermodynamic Permutations for data augmentation, structural amino acid embeddings to model a mutation with a single structure, a protein structure-specific attention-bias mechanism that makes transformers a viable alternative to graph neural networks. We provide training/test splits that mitigate data leakage and ensure proper model evaluation. Furthermore, to examine our data engineering contributions, we fine-tune ESM2 representations (Prostata-IFML) and achieve SOTA for sequence-based models. Notably, Stability Oracle outperforms Prostata-IFML even though it was pretrained on 2000X less proteins and has 548X less parameters. Our framework establishes a path for fine-tuning structure-based transformers to virtually any phenotype, a necessary task for accelerating the development of protein-based biotechnologies.


