2024-11-14 スイス連邦工科大学ローザンヌ校(EPFL)
<関連情報>
- https://actu.epfl.ch/news/simulating-how-fruit-flies-see-smell-and-navigate/
- https://www.nature.com/articles/s41592-024-02497-y
- https://www.nature.com/articles/s41586-024-07939-3
- https://medibio.tiisys.com/97516/
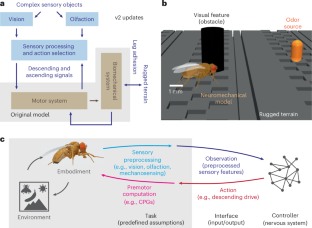
NeuroMechFly v2:ショウジョウバエ成虫の具現化された感覚運動制御のシミュレーション NeuroMechFly v2: simulating embodied sensorimotor control in adult Drosophila
Sibo Wang-Chen,Victor Alfred Stimpfling,Thomas Ka Chung Lam,Pembe Gizem Özdil,Louise Genoud,Femke Hurtak & Pavan Ramdya
Nature Methods Published:12 November 2024
DOI:https://doi.org/10.1038/s41592-024-02497-y
Abstract
Discovering principles underlying the control of animal behavior requires a tight dialogue between experiments and neuromechanical models. Such models have primarily been used to investigate motor control with less emphasis on how the brain and motor systems work together during hierarchical sensorimotor control. NeuroMechFly v2 expands Drosophila neuromechanical modeling by enabling vision, olfaction, ascending motor feedback and complex terrains that can be navigated using leg adhesion. We illustrate its capabilities by constructing biologically inspired controllers that use ascending feedback to perform path integration and head stabilization. After adding vision and olfaction, we train a controller using reinforcement learning to perform a multimodal navigation task. Finally, we illustrate more bio-realistic modeling involving complex odor plume navigation, and fly–fly following using a connectome-constrained visual network. NeuroMechFly can be used to accelerate the discovery of explanatory models of the nervous system and to develop machine learning-based controllers for autonomous artificial agents and robots.
コネクトームに制約されたネットワークがハエの視覚系全体の神経活動を予測する Connectome-constrained networks predict neural activity across the fly visual system
Janne K. Lappalainen,Fabian D. Tschopp,Sridhama Prakhya,Mason McGill,Aljoscha Nern,Kazunori Shinomiya,Shin-ya Takemura,Eyal Gruntman,Jakob H. Macke & Srinivas C. Turaga
Nature Published:11 September 2024
DOI:https://doi.org/10.1038/s41586-024-07939-3
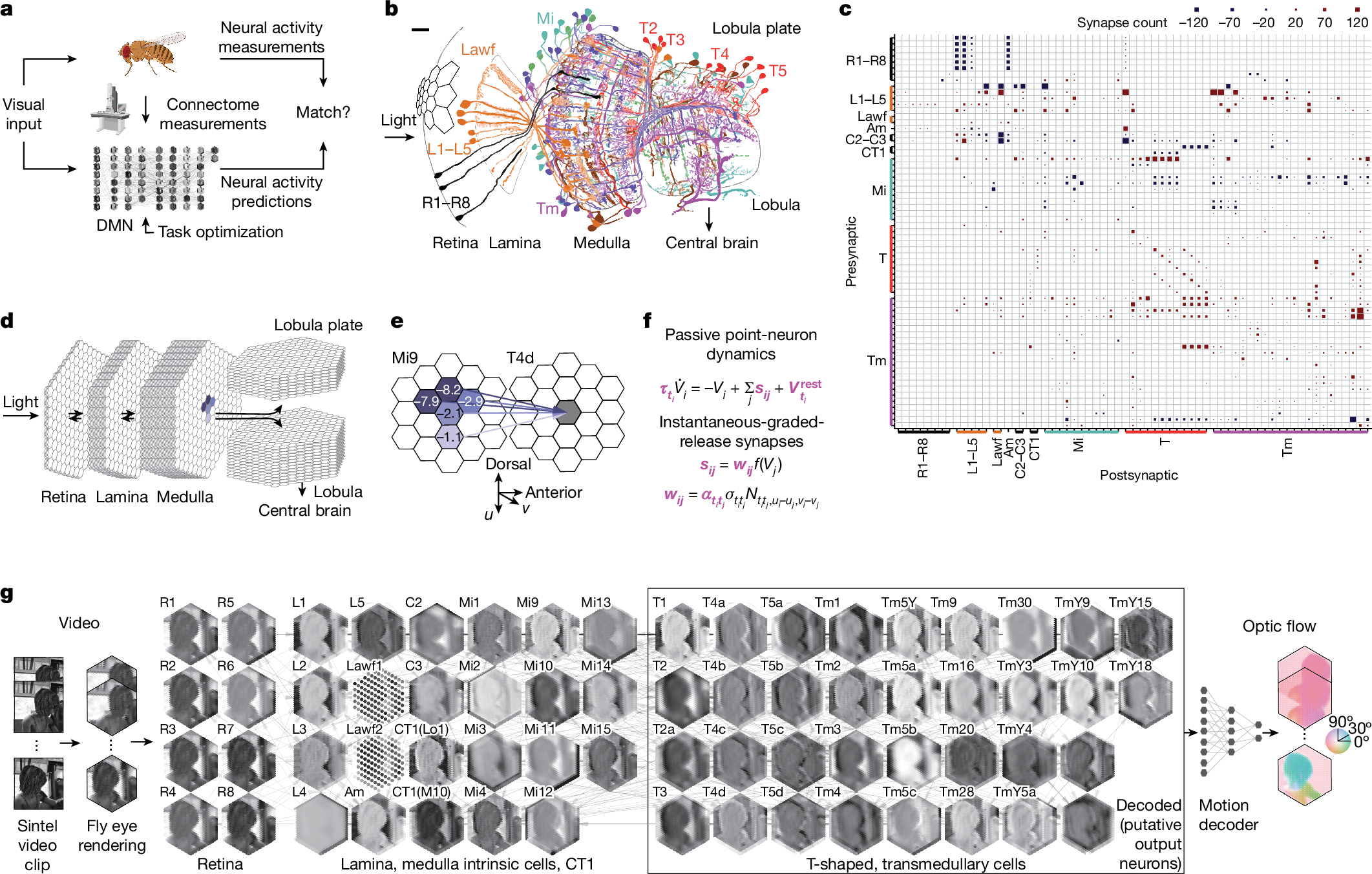
Abstract
We can now measure the connectivity of every neuron in a neural circuit1,2,3,4,5,6,7,8,9, but we cannot measure other biological details, including the dynamical characteristics of each neuron. The degree to which measurements of connectivity alone can inform the understanding of neural computation is an open question10. Here we show that with experimental measurements of only the connectivity of a biological neural network, we can predict the neural activity underlying a specified neural computation. We constructed a model neural network with the experimentally determined connectivity for 64 cell types in the motion pathways of the fruit fly optic lobe1,2,3,4,5 but with unknown parameters for the single-neuron and single-synapse properties. We then optimized the values of these unknown parameters using techniques from deep learning11, to allow the model network to detect visual motion12. Our mechanistic model makes detailed, experimentally testable predictions for each neuron in the connectome. We found that model predictions agreed with experimental measurements of neural activity across 26 studies. Our work demonstrates a strategy for generating detailed hypotheses about the mechanisms of neural circuit function from connectivity measurements. We show that this strategy is more likely to be successful when neurons are sparsely connected—a universally observed feature of biological neural networks across species and brain regions.

